Abstract
Background
Approximately 11% of patients colonized with extended-spectrum beta-lactamase producing Enterobacterales (ESBL-PE) are colonized with more than one ESBL-producing species. We investigated risk factors associated with colonization with multiple ESBL-PE species.
Methods
We performed a case-case–control study at the University Hospital Basel, Switzerland, including hospitalized patients colonized with ESBL-PE between 01/2008 and 12/2018. Patients colonized with multiple species of ESBL-PE during the same hospitalization were assigned to group 1. Group 2 consisted of patients with ESBL-PE and a newly acquired ESBL-PE-species identified during subsequent hospitalization. Controls (i.e., group 3) were patients with only one species of ESBL-PE identified over multiple hospitalizations. Controls were frequency-matched 3:1 to group 2 cases according to time-at-risk (i.e., days between ESBL-PE detection during first and subsequent hospitalizations) to standardize the duration of colonization. ESBL was identified with phenotypic assay and the presence of ESBL genes was confirmed by whole genome sequencing.
Results
Among 1559 inpatients, 154 cases met eligibility criteria (67 in group 1, 22 in group 2, 65 in group 3). International travel within the previous 12 months (OR 12.57, 95% CI 3.48–45.45, p < 0.001) and antibiotic exposure within the previous 3 months (OR 2.96, 95% CI 1.37–6.41, p = 0.006) were independently associated with co-colonization with multiple ESBL-PE species. Admission from another acute-care facility was the only predictor of replacement of one ESBL-PE species with another during subsequent hospitalizations (OR 6.02, 95% CI 1.15–31.49, p = 0.003).
Conclusion
These findings point to strain-related factors being the main drivers of co-colonization with different ESBL-PE and may support stratification of infection prevention and control measures according to ESBL-PE species/strains.
Similar content being viewed by others
Background
Over the past decade, the incidence of extended-spectrum beta-lactamase producing Enterobacterales (ESBL-PE), in particular Escherichia coli and Klebsiella pneumoniae, has increased rapidly worldwide [1], resulting in their classification as serious and critical threats by public health authorities, such as the Centers for Disease Prevention and Control (CDC) and the World Health Organization (WHO) [2, 3]. Asymptomatic colonization has been shown to be a primary risk factor for subsequent ESBL-PE infections [4,5,6]. Several patient-related characteristics have become recognized as risk factors for colonization with ESBL-PE, establishing the foundation for the development of prediction tools [7,8,9,10,11,12,13,14].
Previous investigations have demonstrated that colonization with multiple species of ESBL-PE occur in up to 11% of patients colonized with ESBL-PE [8, 15, 16]. However, data on risk factors and outcomes of co-colonization with different ESBL-PE species are incomplete. Whether patients colonized or infected with multiple ESBL-PE species over time acquire a new ESBL-PE strain or whether their incident ESBL-PE species horizontally transferred plasmids harboring ESBL genes to other colonizing Enterobacterales species remains largely unknown. Understanding this fundamental question will provide insights on the evolving epidemic of ESBL-PE and will inform future infection prevention and antibiotic stewardship interventions to interrupt this pathway. We sought to evaluate patient-related characteristics and exposures associated with colonization with multiple rather than single ESBL-PE species and to identify associated ESBL-gene types to gain further epidemiological insights.
Methods
Setting and participants
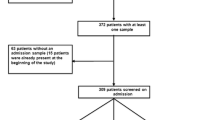
We conducted a retrospective observational case-case–control study at the University Hospital Basel (USB), a tertiary care academic centre admitting over 30,000 patients per year. Patients aged ≥ 18 years and hospitalized from January 2008 until December 2018 with ESBL-PE (as defined below) identified in any clinical or surveillance culture during their hospital-stay were eligible for study inclusion. Herein, “colonization” refers to identification of an ESBL-PE in either a clinical or surveillance culture. Eligible patients and bacterial strains were identified by systematically screening the electronic database of the Clinical Bacteriology and Mycology Laboratory. Strains were accessed via the Clinical Bacteriology and Mycology strain collection. Patients were assigned to the following groups (Fig. 1):
Flowchart of the selection of cases and controls, January 2008–December 2018. aFour patients were eligible for both groups, hence were included in each. b1:3 ratio matching for patients of group 2. Due to one missing control, one case was matched 1:2, resulting in 65 control patients. cafter exclusion of patients of group 1 and 2 as well as patients without a consecutive hospitalization with detection of the same ESBL-PE. ESBL-PE: extended-spectrum beta-lactamase-producing Enterobacterales
Group 1: Patients colonized with multiple species of ESBL-PE (Escherichia coli, Klebsiella species, Proteus spp., Citrobacter spp., Morganella morganii, Serratia marcescens, Enterobacter spp., Pantoea spp.) within the same hospitalization.
Group 2: Patients colonized with ESBL-PE in a first hospitalization and consecutive detection of a different species of ESBL-PE within a following hospitalization during the study period.
Group 3: Patients colonized with only one species of ESBL-PE across multiple hospitalizations during the study period.
Matching between group 2 and group 3 (i.e., the control group) was performed according to time at risk. Time at risk was defined as the number of days between detection of ESBL-PE during the first and subsequent hospitalizations, and was allowed to deviate by a maximum of 25% between cases and controls. These matching criteria were selected to ensure an equitable observation period by providing control patients with the “opportunity” to be colonized with different ESBL-PE species. Three controls were included for each group 2 patient. Risk factors predisposing to case or control status were determined. Additionally, the incidence of ESBL-PE infections and the distribution of ESBL-genes in each group were compared.
This study was approved by the local ethics committee (EKNZ – 2017 00100) and adhered to the Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) guidelines for reporting of observational studies [17].
Data collection
Clinical data were manually extracted from patient’s electronic medical records and entered into a secure REDCap database [18]. Missing data were categorized as negative risk factor. The following variables were collected based on biologic plausibility or their identification in the published literature [19,20,21,22] at the time of each case or control specimen collection date: (1) demographic data; (2) admission and discharge dates and destinations; (3) hospitalization within the previous 12 months, with at least one overnight stay in an acute care or long-term care facility; (4) travel, residence, or hospitalization outside of Switzerland with at least one overnight stay abroad within the previous 12 months; (5) microbiological data (including bacterial genus and species, antibiotic susceptibility data, ESBL status, source of culture, clinical versus surveillance culture, previous history of ESBL-PE colonization or infection); (6) underlying medical conditions on hospital admission based on the Charlson Comorbidity Index (CCI); (7) active open wounds (i.e., diabetic ulcers, decubitus ulcers, or other draining wounds); (8) surgical interventions within six months; (9) indwelling vascular hardware in place for at least 7 days; (10) urinary catheterization (i.e., Foley catheter, intermittent urethral catheterization, ureteral catheters, suprapubic catheterization) within 30 days; (11) history of solid organ or allogenic stem cell transplantation; (12) dialysis; (13) intravenous or oral antibiotic therapy within three months; (14) immunosuppressive therapy (i.e., corticosteroids, calcineurin-inhibitors, mTOR-inhibitors, cytostatics and monoclonal antibodies, or mycophenolates) within 12 months; (15) antacid medications (i.e., proton-pump inhibitors [PPI] or H2-antagonists) within 3 months.
ESBL-PE identification
Stool surveillance specimens were plated onto selective chromogenic agar (chrom ID ESBL, bioMérieux, Marcy-l’Étoile, France). For clinical specimens, bacteria were identified either by MALDI-TOF mass spectrometry (Bruker Daltonics, Bremen, Germany) or by the Vitek 2™ System (bioMérieux, Durham, NC, USA). The Vitek 2™ System was used for susceptibility testing for all isolates. Non-susceptibility to cefpodoxime, ceftriaxone, ceftazidime, or aztreonam was used as a proxy for presumed ESBL production. Phenotypic confirmation of ESBL production was performed using Etest® strips (bioMérieuex, Marcy-l’Etoile, France) or ROSCO disks (Rosco, Taastrup, Denmark). Antibiotic minimal inhibitory concentration (MICs) were interpreted according to EUCAST guidelines (www.eucast.org). Indeterminate results were further evaluated using the Eazyplex Superbug CPE panel (amplex, Gars-Bahnhof, Germany) which include the blaCTX-M-1 and blaCTX-M-9 gene groups. The blaCTX-M-1 and blaCTX-M-9 ESBL groups include the most common ESBL genes identified globally (e.g., blaCTX-M-15, blaCTX-M-14). If these genes were not present, isolates were considered ESBL negative [23].
DNA extraction, library preparation and whole genome sequencing (WGS)
ESBL-PE isolates underwent WGS to identify ESBL genes and to evaluate relatedness of bacterial isolates. Bacteria were grown in blood-agar plates overnight (O/N) and whole bacterial DNA was extracted with the QIAamp DNA Mini kit (QIAGEN) in the QIAcube machine (QIAGEN), according to manufacturer conditions. Genomic libraries were prepared using Nextetra XT protocols (Illumina, San Diego) and WGS was performed using the NextSeq platform (Illumina, San Diego) (read length 2 × 150 bp). Quality control, filtering, and trimming raw sequencing data was performed with the fastp program v.0.20.0 [24]. Antimicrobial resistance genes were predicted directly from the pre-processed FASTQ paired-end reads using the ARIBA tool v.2.14.4 [25] against the ResFinder database [26]. Further classification of the beta-lactamase genes was performed according to the free access lists of the The Galileo AMR database (https://galileoamr.arcbio.com/mara/feature/list), the sequence annotation file of the Digital Multiplex Ligation Assay method validation (https://github.com/manutamminen/dmla), and after exhaustive literature searches.
Statistical analyses
The Fisher’s exact test was used for comparisons of proportions of categorical variables. All continuous variables were found to be abnormally distributed after performing the Shapiro–Wilk test. Therefore, the Mann–Whitney U test was applied to determine medians and interquartile ranges. Univariable analysis with calculation of odds ratios was performed using logistic regression analysis for the non-matched comparisons of patients colonized with multiple ESBL-PE species (group 1) and the control group (group 3). Logistic regression using stepwise forward/backward regression as well as Akaike information criterion (AIC) was performed to identify risk factors independently associated with colonization with multiple ESBL-PE species. Conditional regression analysis was used for comparison of patients with a shift of ESBL-PE (group 2) and matching controls (group 3). p-values ≤ 0.05 were considered significant. STATA version 16.0 (StataCorp, College Station, TX) and R version 3.6.1 (R Foundation for Statistical Computing, Vienna, Austria) were used for statistical analyses.
Results
Characteristics of cases and controls
Among 1559 inpatients colonized with ESBL-PE during the study period, 154 patients met the eligibility criteria (Fig. 1). Group 1 consisted of 67 consecutive patients harbouring multiple ESBL-PE species during a single hospitalization; accounting for 4.3% of all ESBL carriers. Group 2 included 22 cases with new ESBL-PE species identified during subsequent hospitalizations and group 3 consisted of 65 patients with the same ESBL-PE identified during multiple hospitalizations.
Comparisons of baseline characteristics between the three groups are presented in Table 1. The majority of patients were hospitalized in medical wards. Patients were most frequently admitted from home and the distribution of patient age, sex, and burden of comorbidities were similar across groups. Patients belonging to group 1 more frequently reported travelling outside of Switzerland or being hospitalized abroad, and were more likely to be previously exposed to antibiotic treatment, as compared to patients belonging to groups 2 and 3. Group 1 patients were more commonly colonized rather than infected with ESBL-PE as compared to patients belonging to groups 2 and 3. Escherichia coli and Klebsiella pneumoniae were the most frequently detected species of ESBL-PE in each group (Table 1). Length of hospital stay, in hospital death and discharge destination were similar between the three groups (Table 1).
The distribution of different combinations of ESBL-PE within group 1 is shown in Fig. 2, the most prevalent combination being E. coli and K. pneumoniae (69%). Combinations always included either E. coli, K. pneumoniae, or both, along with a less common ESBL-PE species. Within group 2, the shift of species most frequently occurred from E. coli to K. pneumoniae (n = 11, 50%) (Additional file 1: Table S1).
Comparisons across the three groups
Univariable comparisons between groups 1 and 3 revealed travel history, recent hospitalization abroad, and exposure to antibiotic therapy within the prior 3 months to be associated with simultaneous colonization with multiple ESBL-PE species. In an adjusted analysis, including these three variables, travel history and prior antibiotic therapy were independently associated with colonization of multiple ESBL-PE species (OR = 12.57; 95% CI 3.48–45.45, p-value < 0.001 and OR = 2.96; 95% CI 1.37–6.41, p-value = 0.006, respectively; Table 2).
Comparing groups 2 and 3, admission from another acute care facility was the only variable associated with an increased risk of shift of ESBL-PE species (OR 6.02; 95% CI 1.15–31.49, p-value: 0.033); Table 3.
Comparisons between the distribution of ESBL-genes across the three groups
ESBL-strains were available for 153 out of 154 patients (Fig. 1). 277 ESBL-genes were identified and belonged to the following groups: CTX-M-1 group (e.g., blaCTX-M-15, blaCTX-M-1, blaCTX-M-3), CTX-M-8 group (e.g. blaCTX-M-8), CTX-M-9 group (e.g., blaCTX-M-14 and blaCTX-M-27 (a single nucleotide variant of blaCTX-M-14)), and ESBL-variants of blaSHV and blaTEM (Fig. 3a). ESBL-gene-groups in case groups 1 and 2 were similarly distributed, and had higher proportions of blaSHV genes and a lower proportion of blaCTX-M-9 group genes as compared to group 3 (Fig. 3b). Distribution of ESBL genes within the separate species are shown in the Additional file 1: Fig. S1.
Discussion
We found that 4.3% of ESBL-PE carriers were simultaneously colonized with different species of ESBL-PE. Exposure to antibiotic therapy and travel abroad were associated with an increased likelihood of co-colonization with different ESBL-producing species rather than persistent colonization with a single species of ESBL-PE. Admission from another acute care facility was the only variable associated with an increased risk of shift of ESBL-PE species, while antibiotic exposure between hospitalization did not differ between patients with a shift of ESBL-PE species and between patients who remained colonized with a single species of ESBL-PE. Simultaneous or subsequent colonization with different species of ESBL-PE appears to result from either de novo acquisition of ESBL-PE strains or by transmission of ESBL-encoding genes and mobile genetic elements to colonizing non-ESBL-PE strains. The former potentially related to lapses in infection control practices and the later potentially facilitated by antibiotic selection pressure. Our findings support a strong role for the acquisition of novel strains in settings with differing ESBL-PE epidemiology (such as differing geographical regions or healthcare facilities) and suggest that antibiotic selection pressure may facilitate initial colonization, but seems less likely to induce transmission of ESBL-genes to other colonizing Enterobacterales.
Travel to high-endemic areas such as India or South East Asia is a well established risk factor for colonization with ESBL-PE [11], yet co-colonization with different species of ESBL-PE in travelers is presumably low (6.1% among 633 travelers from the Netherlands [11] and 8.6% among travelers from Germany [27] returning with ESBL-PE colonization). The higher frequency of ESBL- E. coli-colonization in returning travelers compared to colonization with other species of ESBL-PE (mainly K. pneumoniae) points to important differences in the epidemiology of these two species of Enterobacterales; E. coli being more likely related to community-acquisition and K. pneumoniae and other ESBL-PE species more likely related to healthcare-associated transmission [28, 29]. Hospitalization abroad did not remain significant in our adjusted analysis as a risk factor for colonization with multiple species of ESBL-PE, likely because of its collinearity with a history of stay outside of Switzerland. Yet, our findings of associations between antibiotic exposures and admission from other healthcare facilities with co-colonization support increased exposure to healthcare services being related with the risk of acquisition of non-E. coli ESBL-PE.
In addition to exposure to settings with differing ESBL-PE epidemiology, antibiotic therapy applying selective pressure has been indicated as a risk factor for colonization with ESBL-PE in various studies [30, 31] and it is not surprising that antibiotic use could foster an environment ripe for carriage of multiple ESBL-PE species. However, our findings did not demonstrate an association between receipt of antibiotic medication between hospitalizations and a shift in species of ESBL-PE. Given the high frequency of antibiotic use, a shift in ESBL-species would likely occur more often, if antibiotic pressure was an important driver of transmission of ESBL-genes within a host.
Across all three case groups investigated in this study, ESBL-genes from the group CTX-M-1 was the most predominant, which corresponds with it being the most widespread group worldwide [32, 33]. Additionally, ESBL-gene-groups in case groups 1 and 2 were similarly distributed, and had higher proportions of SHV genes and a lower proportion of CTX-M-9 group genes as compared to group 3. This difference derives may be related to the higher proportions of K. pneumoniae and from the co-existence of CTX-M-1 and SHV genes in 34.2% and 27.2% of isolates in groups 1 and 2, respectively; while in group 3 only 3.1% of the isolates harboured both gene-groups simultaenously. The presence of the CTX-M-9 group as the second most common EBSL genes in group 3 corresponds to its global prominence [32, 33].
Our study has some important limitations. The retrospective single center study design limits the generalizability of findings to other settings. Travel history is not systematically assessed at hospital admission in our institution and performed mostly in patients with clinical suspicion of travel-acquired infections, patients being repatriated, or patients with a background of migration and its inclusion in our study introduces information bias. A certain number of positive travel histories might have been missed, potentially leading to an overestimation of travel as a risk factor. The limited sample size might have led to our study being underpowered to detect additional risk factors associated with multiple ESBL-PE colonization. Our study occurred in a low ESBL-endemic setting, furthermore a global consensus on active surveillance methods is lacking and our systematic screening strategies for ESBL-PE may vary from other national and international institutions, hence further research is needed to evaluate these findings in regions with a higher prevalence of ESBL-PE.
Conclusions
Co-colonization with different species of ESBL-PE is infrequent and likely to derive from exposure to settings with a differing ESBL-PE epidemiology, as may be encountered in other geographical regions and healthcare settings, further promoted by antibiotic exposure exerting selective colonization-pressure. These results point to specific ESBL-PE strains being the main driver of ongoing ESBL-transmission rather than ongoing host transmission of mobile genetic elements. These results also support the dissemination of ESBL-PE to non-ESBL-PE and support stratification of infection prevention and control measures according to ESBL-PE species/strains. ESBL surveillance frameworks should address potential co-colonization especially in patients with a history of travel abroad or hospitalization at different institutions.
Availability of data and materials
The datasets used and analysed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- AIC:
-
Akaike information criterion
- CCI:
-
Charlson Comorbidity Index
- CDC:
-
Centers for Disease Prevention and Control
- EKNZ:
-
Ethikkommission Nordwest- und Zentralschweiz (ethics commission of northern and central switzerland)
- ESBL:
-
Extended-spectrum beta-lactamase
- ESBL-PE:
-
Extended-spectrum beta-lactamase producing Enterobacterales
- USB:
-
University Hospital Basel
- WGS:
-
Whole genome sequencing
- WHO:
-
World Health Organisation
References
Jernigan JA, Hatfield KM, Wolford H, et al. Multidrug-resistant bacterial infections in U.S. hospitalized patients, 2012–2017. N Engl J Med. 2020;382(14):1309–19.
World Health Organization. Global priority list of antibiotic-resistant bacteria to guide research, discovery and development of new antibiotics. https://www.who.int/medicines/publications/WHO-PPL-Short_Summary_25Feb-ET_NM_WHO.pdf?ua=1. Accessed 09 Dec.
Centers for Disease Control and Prevention. Antibiotic resistance threats in the United States 2019. https://www.cdc.gov/drugresistance/pdf/threats-report/2019-ar-threats-report-508.pdf. Accessed 09 Dec.
Alevizakos M, Karanika S, Detsis M, Mylonakis E. Colonisation with extended-spectrum beta-lactamase-producing Enterobacteriaceae and risk for infection among patients with solid or haematological malignancy: a systematic review and meta-analysis. Int J Antimicrob Agents. 2016;48(6):647–54.
European Centre for Disease Prevention and Control. Surveillance of antimicrobial resistance in Europe 2017: European Centre for Disease Prevention and Control, November 2018. 2018.
Pitout JD, Laupland KB. Extended-spectrum beta-lactamase-producing Enterobacteriaceae: an emerging public-health concern. Lancet Infect Dis. 2008;8(3):159–66.
Lim CJ, Cheng AC, Kennon J, et al. Prevalence of multidrug-resistant organisms and risk factors for carriage in long-term care facilities: a nested case-control study. J Antimicrob Chemother. 2014;69(7):1972–80.
Otter JA, Natale A, Batra R, et al. Individual- and community-level risk factors for ESBL Enterobacteriaceae colonization identified by universal admission screening in London. Clin Microbiol Infect. 2019;25(10):1259–65.
Ny S, Lofmark S, Borjesson S, et al. Community carriage of ESBL-producing Escherichia coli is associated with strains of low pathogenicity: a Swedish nationwide study. J Antimicrob Chemother. 2017;72(2):582–8.
Woerther PL, Andremont A, Kantele A. Travel-acquired ESBL-producing Enterobacteriaceae: impact of colonization at individual and community level. J Travel Med. 2017;24(suppl_1):S29–34.
Arcilla MS, van Hattem JM, Haverkate MR, et al. Import and spread of extended-spectrum beta-lactamase-producing Enterobacteriaceae by international travellers (COMBAT study): a prospective, multicentre cohort study. Lancet Infect Dis. 2017;17(1):78–85.
Kohler P, Fulchini R, Albrich WC, et al. Antibiotic resistance in Swiss nursing homes: analysis of National Surveillance Data over an 11-year period between 2007 and 2017. Antimicrob Resist Infect Control. 2018;7:88.
March A, Aschbacher R, Dhanji H, et al. Colonization of residents and staff of a long-term-care facility and adjacent acute-care hospital geriatric unit by multiresistant bacteria. Clin Microbiol Infect. 2010;16(7):934–44.
Reuland EA, Al Naiemi N, Kaiser AM, et al. Prevalence and risk factors for carriage of ESBL-producing Enterobacteriaceae in Amsterdam. J Antimicrob Chemother. 2016;71(4):1076–82.
Desta K, Woldeamanuel Y, Azazh A, et al. High gastrointestinal colonization rate with extended-spectrum beta-lactamase-producing enterobacteriaceae in hospitalized patients: emergence of carbapenemase-producing K. pneumoniae in Ethiopia. PLoS ONE. 2016;11(8):e0161685.
Pilmis B, Cattoir V, Lecointe D, et al. Carriage of ESBL-producing Enterobacteriaceae in French hospitals: the PORTABLSE study. J Hosp Infect. 2018;98(3):247–52.
von Elm E, Altman DG, Egger M, et al. The Strengthening the Reporting of Observational Studies in Epidemiology (STROBE) statement: guidelines for reporting observational studies. Lancet. 2007;370(9596):1453–7.
Harris PA, Taylor R, Thielke R, Payne J, Gonzalez N, Conde JG. Research electronic data capture (REDCap)—a metadata-driven methodology and workflow process for providing translational research informatics support. J Biomed Inform. 2009;42(2):377–81.
Vock I, Aguilar-Bultet L, Egli A, Tamma PD, Tschudin-Sutter S. Independent, external validation of clinical prediction rules for the identification of extended-spectrum beta-lactamase-producing Enterobacterales, University Hospital Basel, Switzerland, January 2010 to December 2016. Euro Surveill. 2020;25(26).
Vock I, Aguilar-Bultet L, Egli A, Tamma PD, Tschudin-Sutter S. Infections in patients colonized with extended-spectrum beta-lactamase-producing enterobacterales—a retrospective cohort study. Clin Infect Dis. 2021;72(8):1440–43.
Anesi JA, Lautenbach E, Tamma PD, et al. Risk factors for extended-spectrum beta-lactamase-producing enterobacterales bloodstream infection among solid-organ transplant recipients. Clin Infect Dis. 2021;72(6):953–60.
Goodman KE, Lessler J, Harris AD, Milstone AM, Tamma PD. A methodological comparison of risk scores versus decision trees for predicting drug-resistant infections: a case study using extended-spectrum beta-lactamase (ESBL) bacteremia. Infect Control Hosp Epidemiol. 2019;40(4):400–7.
Hinic V, Ziegler J, Straub C, Goldenberger D, Frei R. Extended-spectrum beta-lactamase (ESBL) detection directly from urine samples with the rapid isothermal amplification-based eazyplex(R) SuperBug CRE assay: proof of concept. J Microbiol Methods. 2015;119:203–5.
Chen S, Zhou Y, Chen Y, Gu J. fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics. 2018;34(17):i884–90.
Hunt M, Mather AE, Sanchez-Buso L, et al. ARIBA: rapid antimicrobial resistance genoty** directly from sequencing reads. Microb Genom. 2017;3(10):e000131.
Zankari E, Hasman H, Cosentino S, et al. Identification of acquired antimicrobial resistance genes. J Antimicrob Chemother. 2012;67(11):2640–4.
Lubbert C, Straube L, Stein C, et al. Colonization with extended-spectrum beta-lactamase-producing and carbapenemase-producing Enterobacteriaceae in international travelers returning to Germany. Int J Med Microbiol. 2015;305(1):148–56.
Scheuerman O, Schechner V, Carmeli Y, et al. Comparison of predictors and mortality between bloodstream infections caused by ESBL-producing Escherichia coli and ESBL-producing Klebsiella pneumoniae. Infect Control Hosp Epidemiol. 2018;39(6):660–7.
Freeman JT, Rubin J, McAuliffe GN, et al. Differences in risk-factor profiles between patients with ESBL-producing Escherichia coli and Klebsiella pneumoniae: a multicentre case-case comparison study. Antimicrob Resist Infect Control. 2014;3:27.
Detsis M, Karanika S, Mylonakis E. ICU acquisition rate, risk factors, and clinical significance of digestive tract colonization with extended-spectrum beta-lactamase-producing enterobacteriaceae: a systematic review and meta-analysis. Crit Care Med. 2017;45(4):705–14.
Lee JA, Kang CI, Joo EJ, et al. Epidemiology and clinical features of community-onset bacteremia caused by extended-spectrum beta-lactamase-producing Klebsiella pneumoniae. Microb Drug Resist. 2011;17(2):267–73.
Bevan ER, Jones AM, Hawkey PM. Global epidemiology of CTX-M beta-lactamases: temporal and geographical shifts in genotype. J Antimicrob Chemother. 2017;72(8):2145–55.
Canton R, Gonzalez-Alba JM, Galan JC. CTX-M enzymes: origin and diffusion. Front Microbiol. 2012;3:110.
Acknowledgements
Not applicable.
Funding
This study was funded in part by the University Hospital Basel and the University Basel.
Author information
Authors and Affiliations
Contributions
IV collected patient data, analyzed and interpreted the study data, drafted and revised the manuscript. LA analyzed ESBL-genes, interpreted the study data and revised the manuscript. AE provided data on ESBL-PE identification and revised the manuscript. PT helped interpreting the study data and revised the manuscript. STS conceived the study, analyzed and interpreted the study data and drafted and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was approved by the local ethics committee (Ethikkommission Nordwest- und Zentralschweiz, EKNZ – 2017 00100).
Consent for publication
Not applicable.
Competing interests
All authors declare no conflict of interest relevant to this article.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1.
Supplementary Table 1: Shift of species within group 2. Supplementary Figure 1: Distribution of ESBL genes within separate species.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Vock, I., Aguilar-Bultet, L., Egli, A. et al. Risk factors for colonization with multiple species of extended-spectrum beta-lactamase producing Enterobacterales: a case-case–control study. Antimicrob Resist Infect Control 10, 153 (2021). https://doi.org/10.1186/s13756-021-01018-2
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13756-021-01018-2