Abstract
Integration of single-cell multiomics profiles generated by different single-cell technologies from the same biological sample is still challenging. Previous approaches based on shared features have only provided approximate solutions. Here, we present a novel mathematical solution named bi-order canonical correlation analysis (bi-CCA), which extends the widely used CCA approach to iteratively align the rows and the columns between data matrices. Bi-CCA is generally applicable to combinations of any two single-cell modalities. Validations using co-assayed ground truth data and application to a CAR-NK study and a fetal muscle atlas demonstrate its capability in generating accurate multimodal co-embeddings and discovering cellular identity.
Similar content being viewed by others
Background
Advances in high-throughput single-cell technology such as single-cell RNA-sequencing (scRNA-seq) [1] and mass cytometry [2] have enabled systematic delineation of cell types based on thousands to millions of cells sampled from develo** organisms or patient biopsies [3, 4]. For example, recent application of combinatorial indexing-based technology has generated the transcriptomic and chromatin accessibility profiles of millions of cells in develo** human fetus samples [5]. Rare cell types and complex cellular states, however, remain challenging to discover, which necessitates the development of multiomics technologies to simultaneously measure other cellular features, including DNA methylation [6, 7], chromatin accessibility [8,9,10], and spatial positions [11, 12] in the same cells. Although available single-cell multiomics technologies [10, 13,14,15,16] can profile thousands to millions of cells per experiment, the cost of the experiments is still quite high [17], and the data generated are often of lower throughput than those generated by unimodal technologies. These restrictions necessitate the development of computational approaches that can accurately integrate multiple data matrices generated by different technologies from the same biological samples to acquire an accurate characterization of cellular identity and function.
However, different technologies create data matrices of different rows and columns, which correspond to different sets of cells and different types of features. How to align cells and features simultaneously across matrices is a core computational challenge. When the two sets of cells are sampled uniformly from the same biological sample, it is safe to assume that there exists an optimal alignment of them. However, the search space, whose dimensionality is the product of the numbers of cells (or the numbers of features) in the two sets, is extremely large. To address this challenge, existing computational approaches followed two directions [18]: (1) aligning features empirically before aligning cells [19,20,21,22] and (2) obtaining separate embeddings for each modality, followed by performing unsupervised manifold alignment [23,24,25]. Taking integration of scRNA-seq and single cell assay for transposase accessible chromatin sequencing (scATAC-seq) as an example, the first category of methods require constructing a “gene activity matrix” from scATAC-seq data by counting DNA reads aligned near and within each gene [26]. A successful alignment requires considering both basic proximal regulatory elements and distal regulatory relationship established via other regulatory elements such as enhancers, which are often critical to decipher cell identities [8]. However, current approaches either completely rely on proximal regulatory elements, or infer distal elements from only scATAC-seq data (e.g., Cicero [26]) without integrating with gene expression data. It also substantially simplifies (or loses) multifactorial relations between transcription factors (TF) and target genes [27]. Based on pre-aligned features generated by such empirical rules, Seurat integration (referred to as “Seurat” here after; not to be confused with the weighted nearest neighbor (WNN) approach introduced in Seurat v4 for clustering co-assayed data) applies canonical correlation analysis (CCA) and mutual nearest neighbors (MNNs) to identify cells anchoring the two data matrices [20]; LIGER uses an integrative non-negative matrix factorization (iNMF) to delineate shared and dataset-specific features [22]. Coupled NMF shares similar concept with LIGER [28]; Harmony projects cells onto a shared embedding using principle components analysis (PCA) and removes batch effects iteratively [21]. All these programs suffer from the aforementioned limitations and thereby cannot yield a comprehensive, bi-order gene regulatory network, particularly when chromatin changes are asynchronous from RNA transcriptions in cells undergoing state transitions [29]. The second category of methods such as MATCHER, MMD-MA, UnionCom, SCOT, and Pamona [24, 25, 30,31,32] do not require prior feature alignment. However, they only use intramodal pairwise cell-cell distance information and discard intermodal, trans-acting feature interaction. Thus, they may misalign cell types of similar abundance instead of similar biology, especially rare cell types.
In this study, we develop a novel method called bi-CCA (bi-order canonical correlation analysis) and associated computational tool called bindSC. Bi-CCA learns the optimal alignment among rows and columns (i.e., both cell correspondence and feature interactions) from two data matrices generated by two different experiments. The alignment matrix derived from bi-CCA can thereby be utilized to derive in silico multiomics profiles from aligned cells, which can be used as input to downstream regulatory network inference.
We first assess our method on multimodality integration tasks using benchmarking datasets obtained directly from multiomics technologies, including a novel mouse retinal bipolar cell dataset created by the 10x Genomics Multiome ATAC+RNA kit. Unlike existing integration methods using shared features only, bi-CCA utilizes the full feature information and enables accurate alignment of bipolar cell subtypes between RNA and ATAC data. It also enables discovery of novel cell-type-delineating gene-protein links via integration of RNA and mass cytometry data. We next apply bindSC to two challenging integration tasks. It detects an active immune cell population in the CAR-NK cell products via integration of RNA and mass cytometry data; it resolves mislabeled fetal muscle cells via integration of RNA and ATAC profiles. Bi-CCA is implemented as an open-source R package bindSC available at https://github.com/KChen-lab/bindSC.
Results
Bi-order integration of multi-omics data
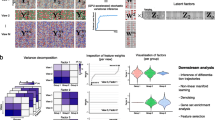
Bi-CCA takes as input two single-cell data matrices (X and Y) generated uniformly from the same cell population by two different technologies (Fig. 1a and Additional file 1: Fig. S1). In most single-cell multi-omics integration tasks, neither the alignment between the cells in X and those in Y, nor the alignment between the features in X and those in Y is known (Additional file 2: Supplementary Note 1). To address this challenge, bi-CCA introduces a modality fusion matrix Z to link X and Y (Fig. 1b). The modality fusion matrix has the same rows as does X and the same columns as does Y. To facilitate the optimization of Z, it is initialized based on prior knowledge linking the two modalities. Taking integration of scRNA-seq and scATAC-seq as an example, the modality fusion matrix can be initialized to the “gene activity matrix” estimated by other programs such as Seurat v3.0. Bi-CCA then iteratively updates Z to find an optimal solution which maximizes the correlation between X and Z and between Y and Z in the latent space simultaneously. Details about this iterative procedure can be found in Methods. In silico simulation experiments using splatter [33] indicate that bi-CCA can robustly align cells and discover meaningful feature interactions from noisy experimental data (Additional file 2: Supplementary Note 2 and Additional file 1: Fig. S2).
Overview of bindSC. a Inputs supported by bindSC. BindSC can integrate two single-cell assays such as transcriptomes, epigenomes, spatial transcriptomes, and proteomes. b Bi-order integration of two modalities (X and Y) with unpaired cells and unmatched features using the bi-CCA approach. In the data matrices, each row represents a gene/locus, and each column represents a cell. Step 1: initializing a modality fusion matrix Z linking the two modalities (Methods). Step 2: matching both cells and features across modalities using CCA. Step 3: updating Z using the obtained cell-cell and feature-feature matching results. Steps 2 and 3 are performed iteratively to optimize Z. c Based on canonical correlation vectors (CCVs) in the derived latent space, bindSC can (1) jointly cluster cells in both modalities to define cell types and (2) transfer labels from one modality to another modality. Association of Z and Y measured in the same cell enables to infer gene-protein and peak-gene regulatory networks. d The integrated multiomics feature profiles enable us to (1) link genes to regulatory elements, (2) map RNA expressions to spatial locations, and (3) delineate cells by both RNA and protein signatures
Bi-CCA outputs canonical correlation vectors (CCVs), which project cells from two datasets onto a shared latent space (hereafter “co-embedding”). Joint clustering, label transfer and network inference can be done in the latent space (Fig. 1c). Moreover, the final modality fusion Z and Y can generate a consensus multiomic profile for cells from Y directly, thus enable (1) characterizing gene and chromatin-accessibility relations from aligned scRNA-seq and scATAC-seq data, (2) associating transcriptomic profiles with proteomic profiles from aligned scRNA-seq and CyTOF data, (3) associating transcriptomic profiles with spatial locations from aligned scRNA-seq and spatial transcriptomic data, and so on (Fig. 1d).
Integration of single-cell RNA-seq and single-cell ATAC-seq data
To examine the utility of bindSC on integrating scRNA-seq and scATAC-seq data, we generated coassayed snRNA-seq and snATAC-seq data using the 10x Genomics Multiome ATAC+RNA kit from an adult mouse retina sample. Mouse retina is heterogeneous, composed of multiple neuronal and non-neuronal cell types [6, 34, 35]. Among them, bipolar cells (BC), which connect photoreceptors (cones and rods) to inner retina, are traditionally dissected into rare subtypes of subtle functional and morphological differences. While high-resolution single-cell transcriptomic profiles of BCs are available [34, Full size image
To evaluate bindSC and three other commonly used methods (Seurat v3.0, LIGER, Harmony) in the task of integrating two independent single-cell dataset, we treated the snRNA and snATAC data as if they were obtained from two different set of cells and tested the ability of these methods in recovering the known pairing. A successful method should project the cells of the same type into the same region in the integration space. As shown in the co-embedding UMAPs (Fig. 2d, e), bindSC successfully achieved that. In the UMAPs generated from the co-embeddings, both the RNA (Fig. 2d) and the ATAC (Fig. 2e) data achieved relatively tight clustering and distributed correspondingly by cell types. We compared cell-ty** accuracy of each method (generated in the respective co-embeddings) with the ground truth. We found that bindSC achieved relatively accurate results (Fig. 2f). In comparison, Seurat v3.0 tended to misalign all cell types to BC1 and had difficulties separating BC8 and BC9. LIGER and Harmony have worse accuracy. These were due partly to the fact that these methods started with gene-based ATAC profiles, which already lost useful information (Fig. 2c).
Because bindSC works with the full ATAC profile, it has the power to better establish the relationship between the RNA and the ATAC features, including potentially distal relationships. To elucidate this point, we calculated the correlation between imputed RNA profiles (i.e., the fusion matrix Z) and the observed RNA profiles. As expected, RNA profiles imputed from gene-based ATAC profiles (at iteration 0) was weakly correlated with the observed RNA profiles (Pearson’s R = 0.1). After 3 iterations, the R value increased to 0.5; meanwhile, the value between imputed and the initial profile decreased to as low as 0.2, indicating the power of associating full peak profiles to genes in a de novo fashion, rather than utilizing reduced profiles (Fig. 2g).
To further examine bindSC’s performance on scenarios where cell populations have imbalanced abundance between two modalities, we generated two datasets: (1) removing 50% of cells in the top five major cell types (i.e., BC1, BC2, BC3, BC4, BC5) in the snATAC data while kee** the snRNA data intact and (2) removing the top five minor cell types (i.e., BC6, BC7, BC8, BC9, BC10) in the snATAC data while kee** the snRNA-seq data intact (Fig. 2h). The label transfer accuracy of bindSC was similar with that from the full paired profiles, indicating bindSC alignment is robust on imbalanced datasets. Again, bindSC had the best performance among all methods in these two scenarios (Fig. 2i–j; Additional file 1: Fig. S4d-e).
We also performed evaluation of several manifold-based methods (SCOT, UnionCom, Pamona, and MMD-MA). They tend to swap entire cell types (Additional file 1: Fig. S4a-b), especially for subtypes with similar abundances, such as BC6 and BC7 (both ~7% abundance; see Additional file 1: Fig. S4a) for SCOT. The map**s, though mathematically plausible, are not biologically sound.
We further examined the 16,944 de novo peak-gene links inferred by bindSC. They can be grouped into 25 clusters. Some of these links were distinct to cell types, while others were shared by multiple cell types (Additional file 1: Fig. S5), indicating potentially a hierarchical regulatory architecture resulting from staged cell lineage differentiation. Specific distal regulatory relations were found in those links, such as Nfib interacting with peaks up to 1Mb away and Car8 interacting with peaks up to 250kb away [37] (Additional file 1: Fig. S6). The integration also enhanced the analysis of correlation between the RNA expression levels of transcription factors (TFs) and their activities inferred from DNA-binding motif enrichment analysis of the ATAC-seq profile (Methods; Additional file 1: Fig. S7).
Overall, our study demonstrated the power of bindSC in generating more accurate in silico multiomics profiles than other existing methods, and the potential in better delineating cell types and associated regulatory signatures.
Integration of single-cell RNA and epitope expression data
Complex interplay exists between mRNAs and proteins [42]. Single-cell proteomic methods such as mass cytometry (CyTOF) [2, 43] measure abundance of a small set of (often 10–50) surface proteins (epitopes) and provide functional quantification of various cell populations. Integrating single-cell RNA and protein data from the same sample can potentially achieve higher resolution characterization and enable discovery of novel cellular states and associated regulatory signatures. This task is challenging because the mRNA and protein expression levels derived from the same genes are not well correlated, due to complex post-transcriptional modifications and technological limitations [44]. CITE-seq [45] performs joint profiling of epitope and mRNA levels in the same cells and can be used to evaluate the results of in silico integration.
We used a CITE-seq dataset consisting of 30,672 human bone marrow cells with a panel of 25 proteins [20]. Unsupervised clustering of the RNA profiles revealed cell types largely consistent with those in the protein profiles, except for some noticeable differences (Fig. 3a, b). CD8+ and CD4+ T cells were partly blended together in the RNA data (ARI = 0.43) but separated clearly in the protein data (ARI = 0.82). On the other hand, conventional dendritic cells (cDC2) were separated from other clusters in the RNA profiles but were intermixed with other cell types in the protein profile. In contrast, the gene expression levels of the 25 RNAs encoding the 25 proteins lacked delineating power and could not yield meaningful classification (ARI = 0.09; Fig. 3c). We randomized the orders of the cells in the RNA matrix and the protein matrix, then tested the ability of each method in generating meaningful co-embeddings and recovering the correct pairing. Seurat v3.0, LIGER and Harmony, which work with only data matrix of 25 homologous features, failed to produce meaningful co-embeddings (Additional file 1: Fig. S8a): the cells from the protein data were well clustered, but those from the RNA data were not meaningfully clustered.
Integrating single-cell RNA with protein data produced by a CITE-seq assay. a–c UMAPs of 30,672 human bone marrow cells based on abundance of the 25 surface proteins (a), RNA expression levels of 3000 highly variable gene (b), and RNA expression levels of the 25 protein-coding genes (c). Labels and dots are colored synchronously by cell type information from the original study. The ARI values are labeled in each panel. d, e UMAPs of the protein (d) and the RNA (e) expression data in the co-embedding generated by bindSC. Each dot in the boxplot denotes one cell type. f Label transfer accuracy of bindSC, Seurat v3.0, LIGER, and Harmony. Each dot in the boxplot denotes one cell type. g Improvement in accuracy of imputed protein level. Each dot represents a protein. X-axis is the Pearson correlation between the ground truth protein level and the RNA level of its coding gene. Y-axis is the Pearson correlation between the ground truth protein level and bindSC imputed protein level. h Comparison of the epitope abundance of CD19, CD14, and CD11c (x-axes) with the RNA expression levels of their coding genes (i.e., CD19, CD14, and ITGAX; y-axes; first row) and with the bindSC imputed protein levels (y-axes; second row). i Gene-protein network inferred from Pearson correlation between genes and bindSC inferred protein levels. A cutoff of 0.55 is used and top five highly correlated genes of each protein are kept
We then tested bindSC on this task. The matrix X was set as the protein matrix, Y the RNA matrix of 3000 highly variable genes, and Z the RNA matrix containing only the 25 protein-homologous genes. Remarkably, the majority of the cells from the two modalities became well aligned in the co-embedding (Fig. 3d, e). Notably, the bulk of CD4+ and CD8+ T cells mixing together in the RNA data became well separated in the co-embedding. We calculated the label transfer accuracy (Methods) between the protein and the RNA cells deriving from the same original cells in the co-embedding. The overall label transfer accuracy for bindSC was significantly higher than those obtained by Seurat, LIGER, and Harmony (Fig. 3f). Overall, the protein levels imputed by bindSC from the entire set of RNAs (i.e., the modality fusion matrix Z) showed consistently higher correlation with the measured epitope levels than the homologous RNA expression levels, indicating meaningful inference of post-transcriptional regulation (Fig. 3g). For example, protein levels for CD19, CD14, and CD11c, markers overexpressing on B cells, monocytes, and DCs, are not highly correlated with the observed RNA expression levels in the same cells (Fig. 3h), however, had much higher correlation with the levels imputed by bindSC from the whole set of RNA expressions. The imputed profile has high correlation with the true protein levels (Pearson’s R = 0.6) and low correlation with the initial gene scores (Pearson’s R < 0.3) (Additional file 1: Fig. S8d), again indicating the power of associating two modalities de novo. We then used the modality fusion matrix Z to infer a gene-protein correlation network (Fig. 3i and Additional file 1: Fig. S9, Methods), in which we see canonical RNA-protein interaction modules centering around CD14 (CD14) and CD79b (CD79B), respectively. Other proteins such as CD19 (CD74, MS4A1, etc.) and CD11a/CD11c (LYZ etc.) have stronger correlation with the RNAs of their upstream or downstream genes, rather than the RNAs of their own coding genes. This result demonstrates the power of bindSC in discovering biologically meaningful regulatory relations and pathways through scRNA-seq and mass cytometry data integration.
Integration of scRNA-seq with CyTOF data revealing activated CAR-NK cells
To further understand the utility of bindSC, we applied it to integrate scRNA-seq and CyTOF data generated from an immunotherapy study. Chimeric antigen receptor (CAR)-transduced natural killer (NK) cells have demonstrated promising efficacy and safety in killing cells in CD19-positive lymphoid tumors [46]. To understand why certain NK cells are more effective than others, we compared the molecular profiles of three groups of NK cells: (1) wildtype non-transduced (NT-NK), (2) transduced with CD19CAR, and (3) transduced with interleukin-15 (IL15).
We obtained scRNA-seq data (1341 cells × 33,538 genes) and CyTOF data (2000 cells × 29 proteins) from the three groups. Clustering the CyTOF and scRNA-seq data by themselves revealed nine and seven clusters (called rClusters and pClusters hereafter), respectively (Fig. 4a, b). After performing bindSC integration, seven integrated clusters (iClusters) were revealed (Fig. 4c, d). Notably, portions of the rClusters R0 and R2, deriving from a subset of CD19CAR NK cells, were reassigned to iCluster 2 (Fig. 4e). Differential expression analysis shows that scRNA-seq cells assigning to iCluster 2 express significantly higher level of inflammation marker TNF, cytokine genes CCL4 and CCL3, and TF genes including JUN and FOS, all indicating activation [47] (Fig. 4g and Additional file 1: Fig. S10). Meanwhile, CyTOF cells assigning to iCluster 2 showed significantly higher levels of 2B4 and DNAM-1 expressions (Fig. 4h), also indicating activation [48]. Importantly, this subset of cells can be identified from neither the scRNA-seq clusters (Fig. 4g), nor the CyTOF clusters alone (Additional file 1: Fig. S11). Thus, integrating scRNA-seq and CyTOF data using bindSC led to the discovery of a subset of highly activated CD19CAR NK cells. This finding may help quantify the therapeutic value of a CAR-NK cell project and reveal mechanisms that can be further leveraged to improve the efficacy of the treatment.
Integration of CyTOF and scRNA data of CD19-CAR NK, IL15 NK, and NT-NK cells. a, b Cells from CD19-CAR NK, IL15 NK, and NT-NK products, clustered independently by CyTOF (a) and scRNA (b) data. There is no correspondence between protein clusters (pClusters) and RNA clusters (rClusters). c, d Integrated clusters (iClusters) after running bindSC on the CyTOF and the scRNA data. CyTOF (a) and scRNA-seq (b) are emphasized, respectively, for better visualization. Cells in the non-emphasized modality are shown in light gray. iCluster 2 is circled out by dashed lines. e Correspondence of iClusters and rClusters. The colors denote the proportion of iCluster in each protein cluster, normalized by each column. The top annotation shows frequencies of three cell groups (cyan: NT-NK, blue: IL15, green: CD19) in each iCluster. f Differentially expressed (Wilcoxon test) genes between iCluster 2 and iClusters 3 and 4. Highlighted genes are known NK cell activation (upregulated). g Gene expression levels of CCL4 and CCL3 in iCluster 2 and rClusters R0 and R2. The p values shown are from the Wilcoxon test. h Protein expression levels of 2B4 and DNAM-1 in iCluster 2, 3, and 5. The p values shown are from the Wilcoxon test
Integration of sci-ATAC-seq and sci-RNA-seq data revealing true identities of rare fetal cells
Bi-CCA alignment may also help identify rare cell populations that are hard to identify in one modality. Recent study used sci-ATAC-seq3 technology to generate the chromatin accessibility profile of ~800,000 human fetal cell atlas from 15 organs [5]. The types of cells in the sci-ATAC-seq data can be annotated by matching clusters with those in the sci-RNA-seq data (Additional file 1: Fig. S12a-b). However, this approach requires good alignment between sci-RNA-seq and sci-ATAC-seq clusters, which is challenging to acquire for rare cell types of limited number of cells. Thus, additional manual review and examination of marker gene expressions are likely required to ensure accurate annotation result. For example, the fetal muscle cell ATAC dataset, consisting of 27,181 cells, has a cluster of cells (3.55% abundance) labeled as unknown (Additional file 1: Fig. S12b), using the above annotation strategy based on gene activity score (ATAC peaks collapsed to genes based on genomic proximity) matrix in the original study. After integrating the sci-ATAC-seq and the sci-RNA-seq data using bindSC, we obtained joint ATAC and RNA profiles (Fig. 5a, b), in which clusters 7 and 8 were annotated as stromal cells (Fig. 5c), different from the previously reported ones (Fig. 5d). We then performed pathway enrichment analysis based on the differentially expressed genes (DEGs) in this cluster (Fig. 5e) and found that these genes are significantly associated with immune (p = 0.003), vascular (p = 0.012), placenta (p = 0.010), and adipose (p = 0.005), indicating that these clusters are highly likely stroma cells surrounding muscle cells. The DEGs are also enriched in biological processes related to extracellular matrix organization (p < 10−4), regulation of exocytosis (p < 10−4) and platelet degranulation (p < 10−4). In comparison, gene activity scores only indicated moderate similarity between clusters 0, 7, and 8, but failed to cluster them together in unsupervised hierarchical clustering (Additional file 1: Fig. S12c-d).
Results for fetal muscle sci-RNA-seq and sci-ATAC-seq data integration. a, b UMAPs generated from bindSC co-embedding of the sci-RNA (a) and the sci-ATAC (b) data. Dots and labels are colored synchronously by cell type. Gray dots represent cells from the other omics (i.e., ATAC cells in a and RNA cells in b). c, d Cell types identified respectively using bindSC (c) and based on gene activity scores in the original publication (d). Clusters 7 and 8, which are classified differently by bindSC, are highlighted by black boxes. e DEGs for each cluster. DEGs specific for clusters 0, 7, and 8 are highlighted by a black box
To examine bindSC’s scalability in large-scale datasets, we created ten benchmark datasets with cells number ranging from 22,552 to 834,424 by resampling cells in the fetal muscle atlas (Additional file 1: Fig. S12e). The block size was set to 1000 for bindSC in each dataset. We obtained the elapsed run time and maximum memory for all the benchmarks using one thread (with a 28-core Intel Skylake CPU@2.6GHz). As expected, bindSC runtime appeared linear to the number of cells, ranging from 4 min for analyzing 23,000 cells to 184 min for 800,000 cells. The maximum memory usage was <10GB in all the datasets, regardless of cell numbers.