Abstract
Background
Hepatitis B virus (HBV)-related hepatocellular carcinoma (HCC) accounts for more than half of total HCC patients in develo** countries. Currently, HBV-related HCC diagnosis and prognosis still lack specific biomarkers. Here, we investigated if PRKRA expression in peripheral blood could be a potential biomarker for the diagnosis/prognosis of HBV-related HCC.
Methods
The expression of PRKRA in HBV-related HCC was firstly analyzed using TCGA and GEO databases. The results were confirmed in a validation cohort including 152 blood samples from 77 healthy controls and 75 HCC patients, 60 of which were infected with HBV. The potential diagnostic and prognostic values of PRKRA were also evaluated by the area under the receiver operator characteristic curve (AUROC) and Kaplan–Meier method, respectively.
Results
PRKRA was significantly upregulated in HCC patients, especially in those with HBV infections. In addition, the combination of PRKRA expression in peripheral blood with serum AFP and CEA levels displayed a better diagnostic performance (AUROC = 0.908, 95% CI 0.844–0.972; p < 0.001). Notably, when serum AFP is less than 200 ng/mL, PRKRA expression demonstrated better diagnostic capability. Furthermore, PRKRA expression levels were associated with expression of EIF2AK2 and inflammatory cytokine genes.
Conclusions
Triple combination testing of blood PRKRA expression, serum AFP and CEA levels could be a noninvasive strategy for diagnosis; and the elevation of PRKRA expression could predicate poor prognosis for HBV-related HCC.
Similar content being viewed by others
Introduction
Hepatocellular carcinoma (HCC) is the sixth most common cancer globally and the second leading cause of cancer-related mortality. Hepatitis B virus (HBV) is one of the most significant risk factors for HCC, especially in HBV epidemic regions [1]. Patients with HBV-associated cirrhosis have a 31-fold increase risk of HCC and 44-fold increase mortality compared to non-cirrhosis patients [2]. In addition, several studies have identified HBV-related factors as crucial predictors of HCC development in patients with chronic hepatitis B, such as serum positive HBV E antigen, high viral load, and viral genotype [2]. Despite an effective vaccine, about 257 million people were infected with HBV [3, 4]. Thus, HBV-related HCC still presents a worldwide threat to public health.
HCC is often diagnosed at advanced stages. The earlier diagnosis was beneficial for the prognosis of HCC since it allowed an early treatment, including surgical resection and a noninvasive ablation. X-ray, biopsy, histopathology, CT/MRI, and serum AFP level are commonly used for HCC diagnosis in clinical practice. Patients with chronic hepatitis were suggested to accept abdominal ultrasound examination together with detection of serum AFP [2, 5]. Nonetheless, current tests with limitations in sensitivity and specificity could result in false positive or inconclusive results. Although advances in cancer care and treatment have improved patient survival, the clinical outcomes of HCC remain dismal, with a less than 50% five-year survival rate after diagnosis [6]. Therefore, it is urgent to identify specific molecules that regulate carcinogenesis and find out potential biomarkers for HCC diagnosis, especially HBV-related HCC.
A double-stranded RNA binding protein (PACT) is encoded by the PRKRA gene. Like argonaute-1 (AGO1), argonaute-2 (AGO2) and TARBP2, PACT belongs to the RNA-induced silencing complex (RISC) [7]. PACT and TARBP2 interact with mammalian DICER to form a complex, which can bind to the ∼70 nt long pre-miRNA “hairpin”, and cleave the loop to produce a double-stranded RNA [8]. Increasing evidence has revealed that these major miRNA pathway components, including PRKRA, were dysregulated in cancers such as epithelial skin cancer and non-small cell lung carcinoma [9, 10]. In addition, PACT was a known cellular protein activator of PKR kinase (also known as EIF2AK2) in a dsRNA‐independent manner in response to cellular stress [11, 12]. PKR could regulate protein translation and trigger the integrative stress response via an eIF2α kinase dependent manner to enhance translation of transcription factors such as ATF4 [13]. Moreover, PACT could enhance the function of retinoic acid-inducible gene I (RIG-I) and induce the expression of type I interferons (IFNs). Both melanoma differentiation-associated gene 5 (MDA5) and laboratory of genetics and physiology 2 (LGP2) were necessary to activate interferon signaling in response to host cell infection by various viruses, including ebola virus (EBOV), influenza A virus, Middle East respiratory syndrome coronavirus, human T-cell leukemia virus 1, mouse hepatitis virus, measles virus, and herpes simplex virus 1 [13, 14].
Recently, PRKRA has been reported to improve oxaliplatin sensitivity in mucinous ovarian cancer cells [15]. The SNP rs2059691 was also associated with increased mRNA expression of PRKRA and worse survival of colorectal cancer patients [16]. However, few studies focused on the link between PRKRA and HCC, especially in HBV-related HCC.
In this study, we firstly found upregulation of PRKRA expression in HBV-related HCC and suggested that the expression of PRKRA in peripheral blood could be a potential biomarker for the diagnosis and prognosis of HBV-related HCC.
Materials and methods
Participants
The PRKRA expression in tumor tissues and clinical information were obtained from The Cancer Genome Atlas (TCGA) dataset (https://portal.gdc.cancer.gov/) and the GEO database (http://www.ncbi.nih.gov/geo). The TCGA cohort contained 371 tumor tissues (HBV-positive group, n = 145; HBV-negative group, n = 226) and 50 tumor-adjacent tissues. The GEO dataset (GSE19665) included 5 pairs of tumor and tumor-adjacent tissues. The validation cohort included 152 blood samples from 75 HCC patients (60 cases of HCC with HBV infection; 64 males and 11 females, mean age 56.5 ± 10.2), and 77 healthy controls (35 males and 42 females, mean age 54.2 ± 9.7) (Additional file 1: Table S1). HCC patients were confirmed by pathology. HCC patients with HBV infection were defined as HBV-related HCC according to serology testing results. The detailed information on liver function and serum tumor biomarkers are described in Additional file 1: Table S1.
Blood RNA extraction and qPCR
Total RNA was isolated from peripheral white blood cells by Trizol reagent (Invitrogen, Carlsbad, USA). RNA was quantified using spectrophotometry (NanoDrop 2000, NanoDrop Technologies, Inc., Wilmington, DE), and the RNA integrity was verified by agarose gel electrophoresis.
The cDNA was synthesized with a reverse-transcription kit with DNAase (Toyobo Co. Ltd., Osaka, Japan). The mRNA expression of PRKRA and EIF2AK2 were determined in triplicates using iTaq™ Universal Supermixes (Bio-Rad, Hercules, CA) with normalization to GAPDH and β-ACTIN. Primer sequences were obtained from Primer Bank (http://pga.mgh.harvard.edu/primerbank) shown in Additional file 1: Table S2.
Statistical analysis
The SPSS software (version 21.0, IBM Inc., Chicago, IL) was used to analyze all data. Comparisons between groups were performed with the Mann–Whitney U test. The prognosis was evaluated by the Kaplan–Meier method and log-rank test. And univariate and multivariate Cox regression models were employed to assess independent prognostic factors. The pairwise deletion method was used to deal with missing data. All the p values were two-sided, and the statistical significance level was at α = 0.05. (*p < 0.05, **p < 0.01, ***p < 0.001.)
Results
PRKRA expression is upregulated in HBV-related HCC
As shown in Fig. 1A, B, the expression of PRKRA was significantly upregulated in HCC tumor tissues, especially in the HBV-positive group (p < 0.001). Moreover, the expression of PRKRA was higher in the advanced TNM-stage group than the early TNM-stage group (p < 0.001) (Fig. 1C). We then compared the mRNA expression levels of PRKRA in tumor tissues and tumor-adjacent tissues from GSE19665 dataset (n = 5), and these results confirmed the increase expression of PRKRA in HCC (p < 0.001) (Fig. 1D).
PRKRA is upregulated in HBV-related HCC. A, B PRKRA mRNA expression in HCC tumor tissues (HBV-positive group, n = 145; HBV-negative group, n = 226) and tumor-adjacent tissues (n = 50). Data were derived from TCGA datasets. C The PRKRA mRNA expression levels in controls and at different TNM stages of HBV-related HCC patients (control, n = 50; stage I, n = 44; stage II, n = 41; stage III + IV, n = 53). D PRKRA mRNA expression in matched tumor and tumor-adjacent tissues from 5 patients. Data were derived from GSE19665 dataset
Higher PRKRA expression levels predict a poor prognosis in HBV-related HCC
Next, we analyzed the association between PRKRA expression and prognosis of HBV-related HCC patients. The patients were divided into PRKRA-low and PRKRA-high groups according to the expression levels of PRKRA in tumors. The Kaplan–Meier analysis indicated that patients in the PRKRA-high group had a poor overall survival (Fig. 2A; log-rank p value < 0.001) and disease-free survival (Fig. 2B; log-rank p value < 0.001) than patients in the PRKRA-low group. Then univariate analysis and multivariate Cox regression models including six parameters (PRKRA expression, AFP levels, sex, age, TNM-stage and pathologic grade) were performed. The results showed that PRKRA expression was correlated with patients' clinical outcomes and was an independent risk factor for HBV-related HCC patients (Hazard ratio = 2.208, 95% CI 1.476–3.304, p < 0.001). Collectively, these results indicate that PRKRA could be a prognosis predictor for HBV-related HCC.
High PRKRA expression levels predict poor prognosis of HBV-related HCC patients. A, B Kaplan–Meier analysis of overall survival (A) and disease-free survival (B) of HBV-related HCC patients based on PRKRA expression (n = 145). The median of PRKRA expression levels was used as the cutoff value for grou** C, D Univariate analysis (C) and multivariate COX regression models (D) including 6 parameters (PRKRA expression, serum AFP level, sex, age, TNM-stage and pathologic grade) were employed to explore risk factors for HBV-related HCC. Symbols indicate Hazard ratio, and bars indicate 95% CIs in forest plots. Uni: Univariate; Mut: multivariate; CI: confidence interval
The diagnostic performance of PRKRA in peripheral blood as a biomarker for HBV-related HCC
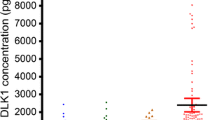
The increased expression of PRKRA in HCC, especially in HBV-related HCC was further confirmed in 152 blood samples from HCC patients and healthy controls (Fig. 3A and Additional file 1: Fig. S1). Then the AUROC was plotted to test whether the PRKRA expression in peripheral blood had diagnostic capacity for HBV-related HCC. As shown in Fig. 3B, PRKRA expression in peripheral blood could distinguish HCC patients from healthy controls with AUROC of 0.713 (95% CI 0.621–0.805; p < 0.001). PRKRA expression combining with serum AFP and CEA showed a much higher AUROC (PRKRA + AFP: 0.880, 95% CI 0.806–0.952; p < 0.001; PRKRA + AFP + CEA: 0.908, 95% CI 0.844–0.972; p < 0.001). Notably, the diagnostic sensitivity of PRKRA expression was 54% (cutoff value = 2.341), while PRKRA expression, serum AFP and CEA served as a combined diagnostic indicator for HBV-related HCC could increase sensitivity to 76%. Besides, PRKRA expression values ≥ 2.341 showed a better diagnostic value (PRKRA: AUROC = 0.952, 95% CI 0.914–0.990; p < 0.001; PRKRA + AFP: AUROC = 0.962, 95% CI 0.920–1.00; p < 0.001; PRKRA + AFP + CEA: AUROC = 0.973, 95% CI 0.934–1.00; p < 0.001). These findings revealed that PRKRA expression, serum AFP and CEA could act as a combined diagnostic indicator for HBV-related HCC.
PRKRA expression in peripheral blood has the potential diagnostic capability for HBV-related HCC. A PRKRA mRNA expression in the validation cohort including 152 blood samples from 77 healthy controls and 75 HCC patients, 60 of which were infected with HBV. B AUROCs for PRKRA, PRKRA + AFP, PRKRA + AFP + CEA in HBV-related patients and healthy controls. C According to the cut-off value (2.341), patients were divided into PRKRA-high and PRKRA-low groups. AUROCs for PRKRA, PRKRA + AFP, PRKRA + AFP + CEA in PRKRA-high group. D In HBV-related patients with AFP ≤ 200 ng/mL and healthy controls, AUROCs for PRKRA, PRKRA + CEA, PRKRA + AFP + CEA
Next, we paid attention to the diagnostic potential of PRKRA in patients with AFP ≤ 200 ng/mL. PRKRA expression levels in peripheral blood (AUROC = 0.746, 95% CI 0.633–0.858; p < 0.001) indicated a better diagnostic capability than serum AFP (AUROC = 0.626, 95% CI 0.477–0.775; p = 0.095) and CEA (AUROC = 0.682, 95% CI 0.558–0.806; p = 0.010) (Fig. 3C). In addition, a combination of PRKRA expression, AFP and CEA could improve the diagnostic capability when serum AFP was at a low level in HBV-related HCC patients. These data suggested that the PRKRA expression in peripheral blood provided the potential diagnostic capability for HBV-related HCC patients.
PRKRA expression levels are associated with EIF2AK2 and inflammatory cytokine genes
To explore the possible reasons for increased PRKRA expression and poor prognosis in HBV-related HCC, we compared the mRNA expression of EIF2AK2 in tumor tissues and blood samples from the HBV-related HCC patients. Similar to the expression pattern of PRKRA, EIF2AK2 was also upregulated both in tumor tissues and peripheral blood samples (Additional file 1: Fig. S2). Increased EIF2AK2 expression levels were also associated with the poor prognosis of HBV-related HCC patients (Additional file 1: Fig. S3).
Pearson correlation analysis was then used to investigate the link between PRKRA, EIF2AK2 and inflammatory cytokine genes (i.e., IL-2, IL-4, IL-6, IL-10, IL-17, IL-22, TNF-α, TGF-α) through a TCGA database including 145 HBV-positive patients. A significant positive correlation between PRKRA expression and EIF2AK2 expression was observed in tissues (r = 0.658, p < 0.001; Fig. 4A) and peripheral blood samples (r = 0.462, p < 0.001; Fig. 4B). Moreover, PRKRA and EIF2AK2 were both positively correlated to the expression of inflammatory cytokine genes, including IL-2 (r = 0.227, p = 0.01; r = 0.241, p < 0.001), IL-4 (r = 0.247, p < 0.001; r = 0.347, p < 0.001), IL-10 (r = 0.227, p = 0.01; r = 0.351, p < 0.001), TNF-α (r = 0.305, p < 0.001; r = 0.446, p < 0.001) and TGF-α (r = 0.389, p < 0.001; r = 0.441, p < 0.001) (Fig. 4 C, D). These results indicated that PRKRA / EIF2AK2 could lead to poor clinical outcomes through activating inflammatory response (Fig. 4F).
PRKRA expression levels are associated with EIF2AK2 and inflammatory cytokines. A, B The positive correlation between PRKRA expression levels and EIF2AK2 expression levels in tissues (A) and peripheral blood samples (B). C, D Heatmaps indicating the associations across PRKRA, EIF2AK2 and inflammatory cytokines genes. Pearson r and p values shown in C, D, respectively. Data were derived from TCGA database. E A schematic illustration of PRKRA upregulation was associated with EIF2AK2 and inflammation, which explain the possible reasons for poor prognosis in HBV-related HCC
Discussion
HBV-related HCC has been a global health problem that accounts for more than half of total HCC patients in develo** countries [17, 18]. In the present study, we found an increase of PRKRA expression in both tumor tissues and peripheral blood samples in HBV-related HCC. The PRKRA expression in peripheral blood combined with serum AFP and CEA displayed a potential diagnostic efficacy with an AUROC of 0.908. To our knowledge, this result is the first time to show the potential diagnostic and prognostic values of PRKRA in HBV-related HCC.
Serum AFP has been the most common biomarker for HCC screening in the last decades, with a sensitivity of 41–65% and a specificity of 80–94% (cutoff value = 20 ng/mL). However, AFP was not secreted by all hepatoma cells, and it might also increase in some patients with cirrhosis or hepatitis [19]. Evidence has shown that nearly half of HCC patients were AFP-negative, especially at an early stage and in small HCCs [19]. Therefore, we focused on the diagnostic efficacy of PRKRA for patients with serum AFP ≤ 200 ng/mL. Interestingly, PRKRA showed a better diagnostic capability than serum AFP.
Furthermore, we also found PRKRA expression was associated with EIF2AK2 and inflammatory cytokine genes, leading to poor prognosis in HBV-related HCC. PRKRA is a known activator of PKR kinase, which plays an essential oncogenic role in HCC. A recent study found that LINC00665, a long intergenic noncoding RNA that physically interacted with PKR, was involved in the NF-κB signaling activation and promoted hepatic cancer progression [20]. PKR activation in stimulated hepatic stellate cells could also promote the development of HCC [21]. In HCC with hepatitis C virus infection, PKR upregulated c-Fos and c-Jun activities to accelerate tumor development [22]. The PKR inhibitor C16 was discovered to block HCC tumor cell growth and angiogenesis in vitro and in vivo through a decrease of growth factors [23]. Mouse xenograft models also confirmed the tumorigenic role of PKR in HepG2 cells by activating STAT3 [24]. Furthermore, PRKRA could bind to the PKR kinase domain and produce interferon and cytokines in virally infected cells [25]. Inflammatory responses to stress can be inhibited by targeting the interaction between PKR and PRKRA [26]. Notably, inflammation was a recognized marker of cancers that substantially contributed to the progression of malignancies, including HBV-related HCC [27]. Consequently, it is reasonable to surmise that the elevated inflammation activated by the increased interaction between PRKRA and EIF2AK2 leads to poor prognosis of HBV-related HCC.
In conclusion, the current study elucidates the elevated expression of PRKRA in HBV-related HCC. The increased PRKRA expression was associated with EIF2AK2 and inflammation, which could explain the possible reasons for poor prognosis in HBV-related HCC. Triple combination testing of blood PRKRA expression and serum AFP and CEA levels could be a noninvasive strategy for the diagnosis and prognosis of HBV-related HCC.
Availability of data and materials
All data are reported in the manuscript.
Abbreviations
- HCC:
-
Hepatocellular carcinoma
- HBV:
-
Hepatitis B virus
- AGO1:
-
Argonaute-1
- AGO2:
-
Argonaute-2
- RISC:
-
RNA-induced silencing complex
- dsRNA:
-
Double-stranded RNA
- RIG-I:
-
Retinoic acid-inducible gene I
- IFNs:
-
Type I interferons
- MDA5:
-
Melanoma differentiation-associated gene 5
- LGP2:
-
Laboratory of genetics and physiology 2
- TCGA:
-
The Cancer Genome Atlas
References
Siegel RL, Miller KD, Jemal A. Cancer statistics, 2020. CA Cancer J Clin. 2020;70(1):7–30. https://doi.org/10.3322/caac.21590.
Sagnelli E, Macera M, Russo A, Coppola N, Sagnelli C. Epidemiological and etiological variations in hepatocellular carcinoma. Infection. 2020;48(1):7–17. https://doi.org/10.1007/s15010-019-01345-y.
An P, Xu J, Yu Y, Winkler CA. Host and viral genetic variatIon in HBV-related hepatocellular carcinoma. Front Genet. 2018;9:261. https://doi.org/10.3389/fgene.2018.00261.
Vallet-Pichard A, Pol S. Review article: immunisation against hepatitis B virus infection and the prevention of hepatocellular carcinoma. Aliment Pharmacol Ther. 2021;53(11):1166–82. https://doi.org/10.1111/apt.16356.
Wu H, Wang MD, Liang L, **ng H, Zhang CW, Shen F, et al. Nanotechnology for hepatocellular carcinoma: from surveillance, diagnosis to management. Small. 2021;17(6): e2005236. https://doi.org/10.1002/smll.202005236.
Li LM, Chen C, Ran RX, Huang JT, Sun HL, Zeng C, et al. Loss of TARBP2 drives the progression of hepatocellular carcinoma via miR-145-SERPINE1 axis. Front Oncol. 2021;11: 620912. https://doi.org/10.3389/fonc.2021.620912.
Galka-Marciniak P, Urbanek-Trzeciak MO, Nawrocka PM, Kozlowski P. A pan-cancer atlas of somatic mutations in miRNA biogenesis genes. Nucleic Acids Res. 2021;49(2):601–20. https://doi.org/10.1093/nar/gkaa1223.
Pullagura SRN, Buaas B, Gray N, Krening LC, Srivastava A, Braun RE. Functional redundancy of DICER cofactors TARBP2 and PRKRA during murine embryogenesis does not involve miRNA biogenesis. Genetics. 2018;208(4):1513–22. https://doi.org/10.1534/genetics.118.300791.
Sand M, Skrygan M, Georgas D, Arenz C, Gambichler T, Sand D, et al. Expression levels of the microRNA maturing microprocessor complex component DGCR8 and the RNA-induced silencing complex (RISC) components argonaute-1, argonaute-2, PACT, TARBP1, and TARBP2 in epithelial skin cancer. Mol Carcinog. 2012;51(11):916–22. https://doi.org/10.1002/mc.20861.
Chiosea S, Jelezcova E, Chandran U, Luo J, Mantha G, Sobol RW, et al. Overexpression of Dicer in precursor lesions of lung adenocarcinoma. Cancer Res. 2007;67(5):2345–50. https://doi.org/10.1158/0008-5472.CAN-06-3533.
Kuipers DJS, Mandemakers W, Lu CS, Olgiati S, Breedveld GJ, Fevga C, et al. EIF2AK2 missense variants associated with early onset generalized dystonia. Ann Neurol. 2021;89(3):485–97. https://doi.org/10.1002/ana.25973.
Shao J, Huang Q, Liu X, Di D, Liang Y, Ly H. Arenaviral nucleoproteins suppress PACT-induced augmentation of RIG-I function to inhibit type I interferon production. J Virol. 2018;92(13): e00482-18. https://doi.org/10.1128/JVI.00482-18.
Dabo S, Maillard P, Collados Rodriguez M, Hansen MD, Mazouz S, Bigot DJ, et al. Inhibition of the inflammatory response to stress by targeting interaction between PKR and its cellular activator PACT. Sci Rep. 2017;7(1):16129. https://doi.org/10.1038/s41598-017-16089-8.
Chukwurah E, Farabaugh KT, Guan BJ, Ramakrishnan P, Hatzoglou M. A tale of two proteins: PACT and PKR and their roles in inflammation. FEBS J. 2021. https://doi.org/10.1111/febs.15691.
Hisamatsu T, McGuire M, Wu SY, Rupaimoole R, Pradeep S, Bayraktar E, et al. PRKRA/PACT expression promotes chemoresistance of mucinous ovarian cancer. Mol Cancer Ther. 2019;18(1):162–72. https://doi.org/10.1158/1535-7163.MCT-17-1050.
Mullany LE, Herrick JS, Wolff RK, Slattery ML. Single nucleotide polymorphisms within MicroRNAs, MicroRNA targets, and MicroRNA biogenesis genes and their impact on colorectal cancer survival. Genes Chromosomes Cancer. 2017;56(4):285–95. https://doi.org/10.1002/gcc.22434.
Lim CJ, Lee YH, Pan L, Lai L, Chua C, Wasser M, et al. Multidimensional analyses reveal distinct immune microenvironment in hepatitis B virus-related hepatocellular carcinoma. Gut. 2019;68(5):916–27. https://doi.org/10.1136/gutjnl-2018-316510.
Zhang H, Chen X, Zhang J, Wang X, Chen H, Liu L, et al. Long noncoding RNAs in HBVrelated hepatocellular carcinoma (Review). Int J Oncol. 2020;56(1):18–32. https://doi.org/10.3892/ijo.2019.4909.
Wang T, Zhang KH. New blood biomarkers for the diagnosis of AFP-negative hepatocellular carcinoma. Front Oncol. 2020;10:1316. https://doi.org/10.3389/fonc.2020.01316.
Ding J, Zhao J, Huan L, Liu Y, Qiao Y, Wang Z, et al. Inflammation-induced long intergenic noncoding RNA (LINC00665) increases malignancy through activating the double-stranded RNA-activated protein kinase/nuclear factor kappa B pathway in hepatocellular carcinoma. Hepatology. 2020;72(5):1666–81. https://doi.org/10.1002/hep.31195.
Imai Y, Yoshida O, Watanabe T, Yukimoto A, Koizumi Y, Ikeda Y, et al. Stimulated hepatic stellate cell promotes progression of hepatocellular carcinoma due to protein kinase R activation. PLoS ONE. 2019;14(2): e0212589. https://doi.org/10.1371/journal.pone.0212589.
Watanabe T, Hiasa Y, Tokumoto Y, Hirooka M, Abe M, Ikeda Y, et al. Protein kinase R modulates c-Fos and c-Jun signaling to promote proliferation of hepatocellular carcinoma with hepatitis C virus infection. PLoS ONE. 2013;8(7): e67750. https://doi.org/10.1371/journal.pone.0067750.
Watanabe T, Ninomiya H, Saitou T, Takanezawa S, Yamamoto S, Imai Y, et al. Therapeutic effects of the PKR inhibitor C16 suppressing tumor proliferation and angiogenesis in hepatocellular carcinoma in vitro and in vivo. Sci Rep. 2020;10(1):5133. https://doi.org/10.1038/s41598-020-61579-x.
Wang X, Dong JH, Zhang WZ, Leng JJ, Cai SW, Chen MY, et al. Double stranded RNA-dependent protein kinase promotes the tumorigenic phenotype in HepG2 hepatocellular carcinoma cells by activating STAT3. Oncol Lett. 2014;8(6):2762–8. https://doi.org/10.3892/ol.2014.2560.
Nakamura M, Kanda T, Sasaki R, Haga Y, Jiang X, Wu S, et al. MicroRNA-122 inhibits the production of inflammatory cytokines by targeting the PKR activator PACT in human hepatic stellate cells. PLoS ONE. 2015;10(12): e0144295. https://doi.org/10.1371/journal.pone.0144295.
Diakos CI, Charles KA, McMillan DC, Clarke SJ. Cancer-related inflammation and treatment effectiveness. Lancet Oncol. 2014;15(11):e493-503. https://doi.org/10.1016/S1470-2045(14)70263-3.
Levrero M, Zucman-Rossi J. Mechanisms of HBV-induced hepatocellular carcinoma. J Hepatol. 2016;64(1 Suppl):S84–101. https://doi.org/10.1016/j.jhep.2016.02.021.
Acknowledgements
The authors would like to thank Dr. Yidan Zhou for her contribution to language polishing.
Funding
This work was supported by the National Natural Science Foundation of China (Grant Nos. 81772276) (to Song-Mei Liu) and Hubei Provincial Natural Science Fund for Creative Research Groups (No. 2019CFA018) (to Song-Mei Liu).
Author information
Authors and Affiliations
Contributions
Y-MH, experimental studies and drafting manuscript; R-XR, sample collection, data analysis and revising manuscript; C-QY, literature search and statistical analysis; S-ML, conceptualization, reviewing manuscript and financial support. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
This study was performed following the Helsinki Declaration and approved by the ethics committee of Zhongnan Hospital of Wuhan University (approval number: 2017058).
Consent for publication
Not applicable.
Competing interests
The authors have declared that no competing interest exists.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1
. Supplementary Table S1. Clinical characteristics of 152 blood samples from HCC patients and healthy controls. Supplementary Table S2. Primers for qRT-PCR used in this study. Supplementary Figure S1. PRKRA is up-regulated in HCC compared with non-HCC and healthy controls. Supplementary Figure S2. EIF2AK2 is up-regulated in HBV-related HCC. Supplementary Figure S3. Higher EIF2AK2 expression levels are associated with a poor prognosis of HBV-related HCC.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Hu, YM., Ran, R., Yang, C. et al. The diagnostic and prognostic implications of PRKRA expression in HBV-related hepatocellular carcinoma. Infect Agents Cancer 17, 34 (2022). https://doi.org/10.1186/s13027-022-00430-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s13027-022-00430-6