Abstract
Background
Previous clinical trials have demonstrated the potential efficacy of poly (ADP-ribose) polymerase (PARP) inhibitors (PARPis) in patients with cancer involving homologous recombination repair (HRR) gene-mutation. Moreover, HRR gene-mutated cancers are effectively treated with immune checkpoint inhibitors (ICIs) with the increase in tumor mutation burden. We have proposed to conduct a multicenter, single-arm phase II trial (IMAGENE trial) for evaluating the efficacy and safety of niraparib (PARPi) plus programmed cell death-1 inhibitor combination therapy in patients with HRR gene-mutated cancers who are refractory to ICIs therapy using a next generation sequencing-based circulating tumor DNA (ctDNA) and tumor tissue analysis.
Methods
Key eligibility criteria for this trial includes HRR gene-mutated tumor determined by any cancer gene tests; progression after previous ICI treatment; and Eastern Cooperative Oncology Group Performance Status ≤ 1. The primary endpoint is the confirmed objective response rate (ORR) in all patients. The secondary endpoints include the confirmed ORR in patients with HRR gene-mutation of ctDNA using the Caris Assure (CARIS, USA). The target sample size of the IMAGENE trial is 57 patients. Biomarker analyses will be performed in parallel using the Caris Assure, proteome analysis, and T cell repertoire analysis to reveal tumor immunosurveillance in peripheral blood.
Expected outcome
Our trial aims to confirm the clinical benefit of PARPi plus ICI combination therapy in ICI-resistant patients. Furthermore, through translational research, our trial will shed light on which patients would benefit from the targeted combination therapy for patients with HRR gene-mutated tumor even after the failure of ICIs.
Trial registration
The IMAGENE trial: jRCT, Clinical trial no.: jRCT2051210120, Registered date: November 9, 2021.
Similar content being viewed by others
Background
Homologous recombination (HR) is a multifactorial process in DNA repair involving the repair of double strand brakes (DSB) generated at the broken replication forks [1]. In particular, HR repair (HRR) genes such as BRCA1 and BRCA2 play a critical role in carrying out successful HR. In normal cells, DSBs are repaired by the HRR mechanism to prevent cell death. By contrast, in cells with HRR dysfunction (HR deficiency: HRD), the damage DNA accumulates because of unsuccessful DSB, eventually leading to cell death [2]. These cells rely on DNA single-strand break repair such as base exchange repair (BER) mechanisms, including Poly (ADP-ribose) polymerase (PARP), an enzyme required for BER to maintain genomic stability [3]. PARP inhibitors (PARPis) induce synthetic lethality in cells with HRD and consequently exhibit antitumor effects. Taking this effect into account, PARPis have undergone clinical approval for the treatment of ovarian, breast and prostate cancers involving HRR gene mutations [4,5,6,7,8,9,10].
Recently, immune checkpoint inhibitors (ICIs) have been introduced with the aim to improve patient outcomes as evidenced by the encouraging results of several clinical trials [11,12,13,14,15,16]. Interestingly, HRR gene mutations are found in a particular patient subset in many types of cancer, and these patients are thought to have susceptibility to ICIs because HRR gene mutations generate immunogenic cancer-antigens owing to the accumulation of somatic mutations [17]. Indeed, several studies have shown that alterations in DNA damage response and repair genes are associated with response to programmed cell death 1/programmed cell death ligand 1 (PD-1/PD-L1) blockade in patients with several types of cancer [18, 19]. Considering above, we can expect synergistic effects of PARPi plus ICI combination therapy for patients with HRR-gene mutations.
The SCRUM-Japan consortium of the Nationwide Cancer Genome Screening Project with an industry-academia collaboration started the MONSTAR-SCREEN project, which evaluates the presence of circulating tumor DNA (ctDNA) before and after initiating cancer treatments [20, 21]. This ctDNA assay can reveal comprehensive somatic genomic alterations that enable the assessment of predominant spatial and temporal intratumoral heterogeneity with minimal invasiveness [22]. Utilizing this platform, we propose an investigator-initiated trial (IMAGENE trial) of niraparib (PARPi) and pembrolizumab or nivolumab combination therapy for HRR gene-mutated solid cancer based on the findings of ctDNA as well as tissue genome screening in eight institutions (sample size: 57, enrollment: 1 years and 8 months).
For personalized immunotherapy, we believe this tumor-agnostic trial would create scientific breakthrough that may lead to better treatments for patients with HRR-gene mutations.
Methods
Study design and treatment
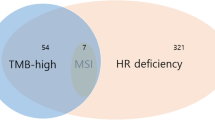
The present trial is a multicenter, single-arm, proof-of-concept (POC), phase II study. aimed to evaluate the efficacy and safety of combination therapy of niraparib plus nivolumab or pembrolizumab (PD-1 inhibitor) in patients with solid cancer involving HRR gene mutations. This study is conducted as a tumor-agnostic trial for patients with metastatic urothelial carcinoma (mUC), metastatic renal cell carcinoma (mRCC), metastatic gastric cancer (mGC), metastatic esophageal cancer (mEC), metastatic head and neck cancer (mHNC), and metastatic melanoma (mMC). HRR gene mutations were determined by tumor tissue analysis including FoundationOne CDx, OncoGuide NCC oncopanel, and Caris Molecular Profiling (Fig. 1). In this study, FoundationOne liquid CDx, Guardant 360, and Caris Assure are also used to screen patients harboring HRR gene mutations using ctDNA tests in blood samples (Fig. 1). The HRR genes are defined as BRCA1, BRCA2, ATM, CDK12, CHEK2, PALB2, BARD1, BRIP1, CHEK1, FANCL, RAD51B, RAD51C, RAD51D, and RAD54L. This trial has been registered in the Japan Registry of Clinical Trials (jRCT2051210120).
Overall trial design. As the translational research, liquid biopsies for the IMAGENE trial will be performed at baseline, cycle 2, and after discontinuation of protocol treatment. C2; Cycle 2, mEC; metastatic esophageal cancer, mGC; metastatic gastric cancer, mHNC; metastatic head and neck cancer, mMC; metastatic melanoma, mRCC; metastatic renal cell carcinoma, mUC; metastatic urothelial carcinoma, PD-1; programmed cell death-1, PD-L1; programmed cell death ligand-1, TCR; T cell receptor
Patients
The eligibility criteria for this trial are presented in Table 1. Key eligibility criteria include the presence of HRR gene mutated tumors determined by an analysis of the tumor tissue or blood sample; tumor progression after treatment with a previous PD-1 or PD-L1 inhibitor; and Eastern Cooperative Oncology Group Performance Status score of ≤ 1. All patients are enrolled in the molecular profiling study, SCRUM-Japan MONSTAR-SCREEN-2 (UMIN000043899), which incorporate a tissue (Caris Molecular Profiling) and plasma multiomics approach (Caris Assure) based on artificial intelligence. Enrollment has started on April 2022 and will be completed on August 2023.
In addition to this clinical trial, patients who do not meet all inclusion criteria, or those meet any of the exclusion criteria are enrolled in the natural history follow-up cohort and will be observed on follow up to collect information on anti-tumor treatment, post-treatment and survival every 3 months as a historical control (SCRUM-Japan registry, UMIN000028058).
Treatments
Patients with mRCC, mGC, mEC, mHNC, and mMC will be treated with niraparib 200 mg daily and nivolumab 3 mg/kg every 2 weeks, whereas those with mUC will be treated with niraparib 200 mg daily and pembrolizumab 3 mg/kg every 3 weeks. Patients will receive therapy until reaching either disease progression or unacceptable toxicity. The dose of niraparib can be reduced to 200 mg daily according to adverse events during the treatment.
Endpoints and statistical analysis
The primary endpoint is the objective response rate (ORR) confirmed by the investigators' assessment in patients with HRR genes mutation determined by any cancer gene panel. The secondary endpoints include the confirmed ORR in patients with HRR gene mutations in ctDNA determined by Caris Assure, progression-free survival (PFS), overall survival (OS), disease control rate (DCR), duration of response (DOR), time to treatment failure (TTF), and change rate of tumor size, as determined by the investigators' assessment, and the incidence of adverse events (AEs). Efficacy will be evaluated according to Response Evaluation Criteria in Solid Tumors (RECIST) V.1.1 using computed tomography every 6 weeks until the end of cycle 12 (nivolumab) or 8 (pembrolizumab), and thereafter every 8 weeks. AEs are assessed according to the Common Terminology Criteria for Adverse Events V.4.0 before the administration of the investigational drug on the day of administration. In this trial, the threshold of the ORR has been set to 16%, and the expected rate to 31%, as niraparib combined with ICIs based on the results of previous clinical trials of ICIs. Under this assumption, with a one-sided significance level of 5% and a power of 80%, the sample size required was calculated to be 57. The sample size for each tumor was set at 15 for mUC, 4 for mRCC, 15 for mGC, 8 for mEC, 13 for mHNC and 2 for mMC, based on feasibility and the expected proportion of participants.
All analysis will be based on the intention-to-treat principle. The baseline characteristics will be described as the mean and standard deviation, or median and quantiles (for continuous variables), or proportion (for categorical variables). The binomial test will be used to analyze the primary endpoint ORR and a threshold, and the Clopper-Pearson method will use the 95% confidence interval for ORR and DOR. Estimates of PFS, DCR, DOR, TTF, and OS will be used with the Kaplan–Meier method.
Biomarker analyses and translational research
ctDNA analysis using a targeted next-generation sequencing (Caris Assure) will be performed at baseline, Cycle 2, and after treatment discontinuation. We will also perform three types of analysis (namely, PD-L1 expression analysis, next-generation sequencing-based T cell repertoire analysis, and quantitative proteomics analysis) to investigate predictive biomarkers of response to this combination therapy. First, the expression level of PD-L1, a well-known biomarker of the clinical response to ICIs in cancer tissues, will be evaluated. Second, we will perform T cell repertoire analysis based on next-generation sequencing to determine whether T cell receptor (TCR) diversity in the peripheral blood can serve as a useful indicator of response to combination therapy. Third, we will perform a mass spectrometry-based quantitative proteomic analysis using plasma to identify candidate proteins as potential predictive biomarkers.
Discussion
The introduction of next-generation sequencing technologies and large-scale tumor molecular profiling tests have revolutionized the field of precision oncology [23]. Precision oncology has already transformed cancer treatment for both common and rare malignancies that are targeted by specific therapies to improve clinical outcomes in patients. HRR gene-mutated tumor are one of the rare fractions across cancers, which are reported to be approximately 3% – 20% [24,25,26,27,31]. Considering that ctDNA may offer clinical advantages for assessing tumor heterogeneity associated with acquired resistance, our trial could contribute to the critical assessment for further use of ctDNA genoty** clinical trials.
Third, our trial results will help perform translational research to decipher the real benefit from PARPi plus ICI combination therapy and could clarify the dynamics of tumor immunosurveillance in both tissues and peripheral blood in our setting. We previously reported that peripheral TCR analysis and early ctDNA dynamics could predict clinical response to ICIs [32]. Franz et al. reported that the clinical utility of proteomic analysis may clarify the responders to PAPRi [33]. Given the additional effect of PAPRi for modulating the tumor microenvironment and cancer-specific T cells, our combined novel analysis could identify the subgroup that shows a good response to this combination therapy.
In conclusion, to the best of our knowledge, the IMAGENE trial is the first phase II study to evaluate the efficacy, safety and POC of combination therapy with niraparib and ICI in patients with HRR gene-mutated tumor. For precision oncology, these findings will shed light on the potential value of targeted combination therapy for patients with HRR gene-mutated tumors even after the failure of ICIs treatment.
Availability of data and materials
Not applicable.
Abbreviations
- CTCAE:
-
Common Terminology Criteria for Adverse Event
- ICI:
-
Immune checkpoint inhibitor
- ORR:
-
Objective response rate
- OS:
-
Overall survival
- PARPi:
-
Poly (ADP-ribose) polymerase inhibitor
- PD-1:
-
Programmed cell death-1
- PD-L1:
-
Programmed cell death ligand-1
- PFS:
-
Progression-free survival
References
Murai J, Pommier Y. PARP trap** beyond homologous recombination and platinum sensitivity in cancers. Ann Rev Cancer Biol. 2019;3:131–50. https://doi.org/10.1146/annurev-cancerbio-030518-055914.
Nguyen L, Martens JWM, Van Hoeck A, Cuppen E. Pan-cancer landscape of homologous recombination deficiency. Nat Commun. 2020;11:5584. https://doi.org/10.1038/s41467-020-19406-4.
Thomas A, Murai J, Pommier Y. The evolving landscape of predictive biomarkers of response to PARP inhibitors. J Clin Invest. 2018;128:1727–30. https://doi.org/10.1172/jci120388.
de Bono J, Mateo J, Fizazi K, Saad F, Shore N, Sandhu S, et al. Olaparib for metastatic castration-resistant prostate cancer. N Engl J Med. 2020;382:2091–102. https://doi.org/10.1056/NEJMoa1911440.
Ledermann J, Harter P, Gourley C, Friedlander M, Vergote I, Rustin G, et al. Olaparib maintenance therapy in platinum-sensitive relapsed ovarian cancer. N Engl J Med. 2012;366:1382–92. https://doi.org/10.1056/NEJMoa1105535.
Mirza MR, Monk BJ, Herrstedt J, Oza AM, Mahner S, Redondo A, et al. Niraparib maintenance therapy in platinum-sensitive, recurrent ovarian cancer. N Engl J Med. 2016;375:2154–64. https://doi.org/10.1056/NEJMoa1611310.
Pujade-Lauraine E, Ledermann JA, Selle F, Gebski V, Penson RT, Oza AM, et al. Olaparib tablets as maintenance therapy in patients with platinum-sensitive, relapsed ovarian cancer and a BRCA1/2 mutation (SOLO2/ENGOT-Ov21): a double-blind, randomised, placebo-controlled, phase 3 trial. Lancet Oncol. 2017;18:1274–84. https://doi.org/10.1016/s1470-2045(17)30469-2.
Gonzalez-Martin A, Pothuri B, Vergote I, DePont CR, Graybill W, Mirza MR, et al. Niraparib in patients with newly diagnosed advanced ovarian cancer. N Engl J Med. 2019;381:2391–402. https://doi.org/10.1056/NEJMoa1910962.
Robson M, Im SA, Senkus E, Xu B, Domchek SM, Masuda N, et al. Olaparib for metastatic breast cancer in patients with a germline BRCA mutation. N Engl J Med. 2017;377:523–33. https://doi.org/10.1056/NEJMoa1706450.
Litton JK, Rugo HS, Ettl J, Hurvitz SA, Gonçalves A, Lee KH, et al. Talazoparib in patients with advanced breast cancer and a germline BRCA mutation. N Engl J Med. 2018;379:753–63. https://doi.org/10.1056/NEJMoa1802905.
Motzer RJ, Escudier B, McDermott DF, George S, Hammers HJ, Srinivas S, et al. Nivolumab versus everolimus in advanced renal-cell carcinoma. N Engl J Med. 2015;373:1803–13. https://doi.org/10.1056/NEJMoa1510665.
Bellmunt J, de Wit R, Vaughn DJ, Fradet Y, Lee JL, Fong L, et al. Pembrolizumab as second-line therapy for advanced urothelial carcinoma. N Engl J Med. 2017;376:1015–26. https://doi.org/10.1056/NEJMoa1613683.
Kang YK, Boku N, Satoh T, Ryu MH, Chao Y, Kato K, et al. Nivolumab in patients with advanced gastric or gastro-oesophageal junction cancer refractory to, or intolerant of, at least two previous chemotherapy regimens (ONO-4538-12, ATTRACTION-2): a randomised, double-blind, placebo-controlled, phase 3 trial. Lancet. 2017;390:2461–71. https://doi.org/10.1016/S0140-6736(17)31827-5.
Kato K, Cho BC, Takahashi M, Okada M, Lin CY, Chin K, et al. Nivolumab versus chemotherapy in patients with advanced oesophageal squamous cell carcinoma refractory or intolerant to previous chemotherapy (ATTRACTION-3): a multicentre, randomised, open-label, phase 3 trial. Lancet Oncol. 2019;20:1506–17. https://doi.org/10.1016/S1470-2045(19)30626-6.
Ferris RL, Blumenschein G Jr, Fayette J, Guigay J, Colevas AD, Licitra L, et al. Nivolumab for Recurrent Squamous-Cell Carcinoma of the Head and Neck. N Engl J Med. 2016;375:1856–67. https://doi.org/10.1056/NEJMoa1602252.
Weber JS, D’Angelo SP, Minor D, Hodi FS, Gutzmer R, Neyns B, et al. Nivolumab versus chemotherapy in patients with advanced melanoma who progressed after anti-CTLA-4 treatment (CheckMate 037): a randomised, controlled, open-label, phase 3 trial. Lancet Oncol. 2015;16:375–84. https://doi.org/10.1016/s1470-2045(15)70076-8.
Yarchoan M, Johnson BA 3rd, Lutz ER, Laheru DA, Jaffee EM. Targeting neoantigens to augment antitumour immunity. Nat Rev Cancer. 2017;17:209–22. https://doi.org/10.1038/nrc.2016.154.
Teo MY, Seier K, Ostrovnaya I, Regazzi AM, Kania BE, Moran MM, et al. Alterations in DNA damage response and repair genes as potential marker of clinical benefit from PD-1/PD-L1 blockade in advanced urothelial cancers. J Clin Oncol. 2018;36:1685–94. https://doi.org/10.1200/jco.2017.75.7740.
Labriola MK, Zhu J, Gupta RT, McCall S, Jackson J, Kong EF, et al. Characterization of tumor mutation burden, PD-L1 and DNA repair genes to assess relationship to immune checkpoint inhibitors response in metastatic renal cell carcinoma. J Immunother Cancer. 2020;8:e000319. https://doi.org/10.1136/jitc-2019-000319.
Nakamura Y, Fujisawa T, Taniguchi H, Bando H, Okamoto W, Tsuchihara K, et al. SCRUM-Japan GI-SCREEN and MONSTAR-SCREEN: Path to the realization of biomarker-guided precision oncology in advanced solid tumors. Cancer Sci. 2021. https://doi.org/10.1111/cas.15132.
Taniguchi H, Nakamura Y, Kotani D, Yukami H, Mishima S, Sawada K, et al. CIRCULATE-Japan: Circulating tumor DNA-guided adaptive platform trials to refine adjuvant therapy for colorectal cancer. Cancer Sci. 2021;112:2915–20. https://doi.org/10.1111/cas.14926.
Parikh AR, Leshchiner I, Elagina L, Goyal L, Levovitz C, Siravegna G, et al. Liquid versus tissue biopsy for detecting acquired resistance and tumor heterogeneity in gastrointestinal cancers. Nat Med. 2019;25:1415–21. https://doi.org/10.1038/s41591-019-0561-9.
Malone ER, Oliva M, Sabatini PJB, Stockley TL, Siu LL. Molecular profiling for precision cancer therapies. Genome Med. 2020;12:8. https://doi.org/10.1186/s13073-019-0703-1.
Robertson AG, Kim J, Al-Ahmadie H, Bellmunt J, Guo G, Cherniack AD, et al. Comprehensive molecular characterization of muscle-invasive bladder cancer. Cell. 2017;171(540–56):e25. https://doi.org/10.1016/j.cell.2017.09.007.
Sato Y, Yoshizato T, Shiraishi Y, Maekawa S, Okuno Y, Kamura T, et al. Integrated molecular analysis of clear-cell renal cell carcinoma. Nat Genet. 2013;45:860–7. https://doi.org/10.1038/ng.2699.
Comprehensive molecular characterization of gastric adenocarcinoma. Nature. 2014;513:202–9. https://doi.org/10.1038/nature13480.
Integrated genomic characterization of oesophageal carcinoma. Nature. 2017;541:169–75. https://doi.org/10.1038/nature20805.
Heeke AL, Pishvaian MJ, Lynce F, **u J, Brody JR, Chen WJ, et al. Prevalence of homologous recombination-related gene mutations across multiple cancer types. JCO Precis Oncol. 2018;2018:17. https://doi.org/10.1200/po.17.00286.
Kim KB, Soroceanu L, de Semir D, Millis SZ, Ross J, Vosoughi E, et al. Prevalence of homologous recombination pathway gene mutations in melanoma: rationale for a new targeted therapeutic approach. J Invest Dermatol. 2021;141:2028. https://doi.org/10.1016/j.jid.2021.01.024.
Césaire M, Thariat J, Candéias SM, Stefan D, Saintigny Y, Chevalier F. Combining PARP inhibition, radiation, and immunotherapy: A possible strategy to improve the treatment of cancer? Int J Mol Sci. 2018;19:3793. https://doi.org/10.3390/ijms19123793.
Nakamura Y, Okamoto W, Kato T, Esaki T, Kato K, Komatsu Y, et al. Circulating tumor DNA-guided treatment with pertuzumab plus trastuzumab for HER2-amplified metastatic colorectal cancer: a phase 2 trial. Nat Med. 2021;27:1899–903. https://doi.org/10.1038/s41591-021-01553-w.
Kato T, Kiyotani K, Tomiyama E, Koh Y, Matsushita M, Hayashi Y, et al. Peripheral T cell receptor repertoire features predict durable responses to anti-PD-1 inhibitor monotherapy in advanced renal cell carcinoma. Oncoimmunol. 2021;10:1862948. https://doi.org/10.1080/2162402x.2020.1862948.
Franz A, Coscia F, Shen C, Charaoui L, Mann M, Sander C. Molecular response to PARP1 inhibition in ovarian cancer cells as determined by mass spectrometry based proteomics. J Ovarian Res. 2021;14:140. https://doi.org/10.1186/s13048-021-00886-x.
Acknowledgements
We would like to thank all the patients and their families who participated in this trial; all the co-investigators and site personnel. We also thank the clinical trial office members in the Department of Medical Innovation, Osaka University Hospital (Yoko Kawamura, Kento Asano, and Tomoharu Dohi) and FiveRings Corporation (Harumi Sakata, Chiho Uegaki, Tatsushi Goto, and Hiroshi Miyamoto).
Funding
IMAGENE trial receives financial support from the Japan Agency for Medical Research and Development (AMED; grant Number, 21ck0106656h0001). Takeda Pharmaceutical Co., Ltd. provides investigational drugs as well as financial support for the translational research. The funders had no role in the study design; in data collection, analysis, or interpretation; or in the writing of the report.
Author information
Authors and Affiliations
Contributions
TK and NN participated in the entire coordinating of this trial, designed and prepared the protocol. NM, MS, ME, TO, TA, NS, YY, NT, MO, KN, TH, MN, TK, KN, TF, SO, YN, HB, and TY designed and prepared the protocol. EH is the chief statistician. All authors approved the final version of the manuscript for submission and agreed to be accountable for the contents.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
The IMAGENE trial is conducted in accordance with the Declaration of Helsinki, the Japanese Ethical Guidelines for Medical and Health Research Involving Human Subjects, and the Clinical Trial Acts in Japan. Each trial has been approved by the institutional review board of Osaka University Hospital Certified Review Board.
All patients were required to sign written informed consent.
Registry and the Registration No. of the study/trial: This trial has been registered in the Japan Registry of Clinical Trials (jRCT2051210120).
Consent for publication
Not applicable.
Competing interests
TK reports research funding supports from Takeda and Pfizer. NM reports honoraria from Janssen, Merck biopharma, and Sanofi and research funding supports from Janssen, AstraZeneca, Bayer, Roche, MSD, Taiho, Astellas, Amgen, Eisai, Eli Lilly, PRA Health Science, Takeda, Pfizer, Seagen, Chugai, Abbvie, and Novartis. MS reports honoraria from Janssen, AstraZeneca, and Astellas and research funding support from Daiichi Sankyo. ME reports honoraria from ONO, Takeda, Novartis, Pfizer, Bristol, Janssen, MSD, Merck, AstraZeneca, and Eisai and research funding support from Sanofi, Bayer, Astellas, ONO, and Takeda. TO reports honoraria from Takeda. MO reports honoraria from Bayer, Bristol, Novartis, Ono, Merck, Takeda, MSD, and Pfizer. KN reports honoraria from Ono, Novartis, MSD, and Bristol, and consulting fees from Novartis and MSD. YN reports honoraria from Chugai, Merck Biopharma, and Guardant Health AMEA, and research funding support from Taiho, Chugai, Guardant Health, Genomedia, Daiichi Sankyo, Seagen, and Roche Diagnostics. HB reports honoraria from Taiho, Ono, and Eli Lilly Japan, and research funding support from Ono. TY reports honoraria from Chugai, Merck, Bayer, Ono, and MSD, and research funding from MSD, Daiichi Sankyo, Ono, Taiho, Amgen, Sanofi, Pfizer, Genomedia, Sysmex, Chugai, and Nippon Boehringer Ingelheim. NN reports honoraria from Takeda, Janssen, AstraZeneca, Merck Biopharma, Ono, and Bristol.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Kato, T., Matsubara, N., Shiota, M. et al. IMAGENE trial: multicenter, proof-of-concept, phase II study evaluating the efficacy and safety of combination therapy of niraparib with PD-1 inhibitor in solid cancer patients with homologous recombination repair genes mutation. BMC Cancer 22, 1292 (2022). https://doi.org/10.1186/s12885-022-10398-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12885-022-10398-6