Abstract
Background
Drug resistance is still one of the key causes of death in epithelial ovarian carcinoma (EOC) patients, however there are very few strategies to reverse chemoresistance. Here we try to clarify whether and how miR-9 takes part in the regulation of paclitaxel sensitivity.
Methods
miR-9 expressions in EOC cells and tissues were detected by Realtime PCR. The target of miR-9 was validated through dual luciferase reporter assay and Western Blot. Methylation study, RNAi technique and cytotoxicity assay were used to determine the intrinsic mechanism of miR-9 in paclitaxel sensitivity regulation.
Results
miR-9 is down-regulated in paclitaxel resistant EOC. The patients with lower miR-9, Grade 3, Stage III –IV and suboptimal surgery present shorter survival time. miR-9 and suboptimal surgery are independent prognostic factors of EOC. Modulating miR-9 expression could change paclitaxel sensitivity of EOC cells. CCNG1, validated as a direct target of miR-9, mediates paclitaxel resistance. miR-9-1 and 3 gene hypermethylation would decrease miR-9 expression, while demethylation of miR-9 gene could restore miR-9 expression and improve paclitaxel sensitivity in chemoresistance EOC cells. Furthermore, methylation-associated miR-9 deregulation in EOC cells could be induced by paclitaxel exposure.
Conclusions
Methylation-associated miR-9 down-regulation is probably one of the key mechanisms for paclitaxel resistance in EOC cells, via targeting CCNG1. Our findings may also provide a new potential therapeutic target to reverse paclitaxel resistance in EOC patients.
Similar content being viewed by others
Background
Although improved combination of surgery and chemotherapy is commonly applied for epithelial ovarian carcinoma (EOC), EOC is still the leading cause of death among gynecologic cancer nowadays [1, 2]. Initial response to chemotherapeutic drugs can be achieved in about 70 % EOC patients, but most of patients eventually develop chemoresistance and succumb to their diseases [1, 3]. Numerous evidences showed that chemoresistance is a clinically formidable problem in managing EOC patients. Obviously, reversion of drug resistance would contribute to improve prognosis.
As we know,miRNAs are small noncoding RNAs involved in the initiation and progression of human cancer [4]. Previous studies have suggested that miRNAs can act as oncogenes or tumor suppressors, exerting a key function in tumorigenesis and progression [5, 6]. For instance, miR-9 is under-expressed in gastric cancer, and ectopic expression of miR-9 can influence cell growth and cell cycle [7]. As a highly conserved miRNA found in insects and primates, miR-9 has three independent gene loci: miR-9-1, miR-9-2, and miR-9-3, located at chromosomes 1, 5, and 15, respectively [8]. miR-9 expression can be epigenetically modified. Studies showed that miR-9 deregulation and gene methylation was associated with cancer development and recurrence [9–11]. Heller [12] recently found that miR-9-3 methylation was related to shorter overall survival and disease-free survival of lung squamous cell carcinoma patients. But no study, to our best knowledge, has been reported about the intrinsic relationship between miR-9 deregulation and paclitaxel resistance in cancer research up to today.
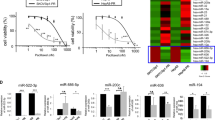
Our previous studies have identified a deregulated miRNA profile in paclitaxel resistant EOC using miRNA microarray and Realtime PCR [Full size image
CCNG1 is one of the targets directly regulated by miR-9
TargetScan database (www.targetscan.org, Released 6.0, Nov 2011) predicted CCNG1 contain the putative miR-9 binding site. Western blot validated that up-regulated miR-9 could inhibit CCNG1 expression in ST30 cells, while down-regulated miR-9 enhanced CCNG1 expression in SKOV3 cells (Fig. 2b, e). Using dual luciferase reporter assay, we found that the relative luciferase activities were significantly reduced in cells transfected with CCNG1 WT- 3′UTR/miR-9 mimic vectors compared with those transfected with CCNG1 WT-3′UTR/miR-9 mimic control (Fig. 2c). Furthermore, Realtime RT-PCR suggested that mRNA expression of CCNG1 in ST30 cells was significantly higher than that in SKOV3 (2.14 fold), which was contrary to the miR-9 expression trends in SKOV3 and ST30 cells (Additional file 2: Figure S1C). These data validate that miR-9 can directly bind to 3′UTR of CCNG1 and CCNG1 is regulated by miR-9.
CCNG1 depletion enhances the paclitaxel sensitivity of EOC cells
CCNG1 was initially identified as a p53-regulated transcript induced by DNA damage [15]. Although its precise role on cellular growth control is still controversial, CCNG1 has been regarded as an oncogene [16, 17]. CCNG1 gene copy number is an independent marker of postsurgical survival in EOC patients who have received chemotherapy with taxanes and platinum compounds [18]. Thus it suggests that CCNG1, the target of miR-9, probably modulates the paclitaxel-sensitivity of EOC. To validate this hypothesis, we knocked down CCNG1 in ST30 cells through transfecting CCNG1 siRNA. The roles of CCNG1 siRNAs were confirmed using realtime RT-PCR and Western blot, and the most effective siRNA was chosen (Additional file 2: Figure S1D, E). IC50 of paclitaxel in ST30 cells was significantly decreased after CCNG1 depletion (Fig. 2f), which confirmed that knockdown of CCNG1 alone would enhance paclitaxel cytotoxicity to EOC cells. Furthermore, when miR-9 inhibitor was transfected into SKOV3 cell accompanied with CCNG1 siRNA, the CCNG1 expression and the affection of miR-9 inhibitor on paclitaxel sensitivity were significantly reversed (Fig. 2e, f). Therefore, the results indicate that CCNG1, as one direct target of miR-9, participates in the regulation of paclitaxel-sensitivity in EOC cells.
miR-9-1 and 3 loci are hypermethylated in paclitaxel resistant EOC cells
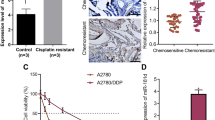
BSP revealed higher frequency of DNA methylation of miR-9-1 and miR-9-3 in ST30 and A2780R cells compared with their parental cells (Fig. 3a-c and Table 2). Again, MSP presented similar results (Fig. 3d). We further detected the methylation status of all three independent miR-9 gene loci in 66 EOC tissues using MSP and found chemo-resistant specific DNA hypermethylation patterns. The miR-9-1 and 3 genes exhibited a significantly DNA hypermethylation status in the chemoresistant tissues compared with chemosensitive tissues, as shown in Table 3. Thus, our data suggest that decreased miR-9 expression might result from DNA hypermethylation in EOC cells.
Methylation status of Hsa-miR-9 genes in EOC cells and their effects on the paclitaxel sensitivity. a. BSP results of the hsa-miR-9-1 CpG island region in two pairs of paclitaxel sensitive and resistant EOC cell lines. 10 clones were sequenced for each cell line. Each circle indicates a CpG dinucleotide, black circle: methylated CpG; open circle: unmethylated CpG. b. BSP results of the hsa-miR-9-2 CpG island region in two pairs of paclitaxel sensitive and resistant EOC cell lines. 10–12 clones were sequenced for each cell line. c. BSP restuls of the hsa-miR-9-3 CpG island region in two pairs of paclitaxel sensitive and resistant EOC cell lines. 11 clones were sequenced for each cell line. d. MSP of three hsa-miR-9 genes in different ovarian carcinoma cell lines. e. Real-time RT-PCR analysis of miR-9 in ST30 cell lines treated with or without DAC. Low miR-9 expression in ST30 cells was restored by DAC (P = 0.000). f. The effect of DAC and miR-9 inhibitor on the paclitaxel sensitivity of ST30 cell lines. The inhibited rates of paclitaxel on ST30 cell lines treated with or without miR-9 inhibitor, DAC combined with miR-9 inhibitor or DAC were assessed by MTS assay and the IC50 values were 3295.54 ± 154.87nM, 2590.36 ± 126.68nM, and 2057.35 ± 13.54nM in turn, compared with control (IC50 = 2898.94 ± 155.75 nM), P = 0.001, 0.056 and 0.035 in turn. The experiments were repeated three times
Regulating miR-9 gene methylation changes chemo-sensitivity of ST30 cells
To confirm the influence of DNA methylation on miR-9 expression and chemo- sensitivity in EOC cells, miR-9 level of EOC cells were detected after cultured with or without DAC, an unspecific demethylation reagent. We found that low miR-9 expression in ST30 cells could be restored by genomic DNA hypomethylation (Fig. 3e). Therefore, Regulating DNA methylation would change the transcriptional activity of miR-9 in paclitaxel resistant EOC cells.
MTS assay showed that the IC50 value of ST30 was significantly increased in miR-9 inhibitor treated group and significantly decreased in DAC treated group, compared with that in miR-9 inhibitor control group. While DAC combined with miR-9 inhibitor group had no significantly effect on IC50 of ST30. (Fig.3f). These results suggest that hypermethylation of miR-9 genes, mainly miR-9-1 and miR-9-3, would down-regulate miR-9 expression of EOC. Consequently, decreased miR-9 expression results in paclitaxel resistance of EOC cells. Thus, miR-9 is probably a potential therapeutic target and ablation of miR-9 hypermethylation status may partly reverse chemo-sensitivity in paclitaxel resistance EOC cells.
Paclitaxel induces decreased miR-9 expression in EOC cells through influencing DNA methylation
SKOV3 and A2780 were exposed to 10uM paclitaxel for 60 min before the drug was washed out. The miR-9 levels decreased after drug exposure in both cell lines, with the peak at 24 h (Fig. 4d), suggesting that the changes in miR-9 expression might be induced by paclitaxel. Since DNA hypermethylation was confirmed in paclitaxel resistant cells, we further determined whether the DNA methlyation status of ovarian carcinoma cell lines was changed after paclitaxel treatment. BSP results (Fig. 4a, c and Table 2) revealed a higher frequency of DNA methylation of miR-9-1 and 3 in both cell lines at 24 h after drug exposure. In addition, DNA methylation of miR-9-2 was also increased after drug exposure, although not significantly (Fig. 4b). Our findings suggest that paclitaxel resistance produced by methylation- associated miR-9 deregulation may be secondary from paclitaxel treatment in EOC cells.
Paclitaxel down-regulates miR-9 expression in EOC cells through influencing DNA methylation. a. BSP results of the hsa-miR-9-1 CpG island region in different ovarian carcinoma cell lines before and after 24 h paclitaxel treatment. 10 clones were sequenced for each cell line. Each circle indicates a CpG dinucleotide, black circle: methylated CpG; open circle: unmethylated CpG. b. BSP results of the hsa-miR-9-2 CpG island region in different ovarian carcinoma cell lines before and after 24 h paclitaxel treatment. 10–12 clones were sequenced for each cell line. c. BSP results of the hsa-miR-9-3 CpG island region in different ovarian carcinoma cell lines before and after 24 h paclitaxel treatment. 10–11 clones were sequenced for each cell line. d. Real-time RT-PCR analysis of miR-9 in SKOV3 and A2780 cell lines treated with paclitaxel at different time. The experiments were repeated three times