Abstract
Background
The occurrence of multidrug-resistant and hypervirulent Klebsiella pneumoniae (MDR-hvKp) worldwide poses a great challenge for public health. Few studies have focused on ST218 MDR-hvKp.
Methods
Retrospective genomic surveillance was conducted at the Peking University Third Hospital from 2017 and clinical information was obtained. To understand genomic and microbiological characteristics, antimicrobial susceptibility testing, plasmid conjugation and stability, biofilm formation, serum killing, growth curves and whole-genome sequencing were performed. We also assessed the clinical and microbiological characteristics of ST218 compared with ST23.
Results
A total of eleven ST218 Kp isolates were included. The most common infection type was lower respiratory tract infection (72.7%, 8/11) in our hospital, whereas ST23 hvKp (72.7%, 8/11) was closely associated with bloodstream infection. Notably, nosocomial infections caused by ST218 (54.5%, 6/11) was slightly higher than ST23 (36.4%, 4/11). All of the ST218 and ST23 strains presented with the virulence genes combination of iucA + iroB + peg344 + rmpA + rmpA2. Interestingly, the virulence score of ST218 was lower than ST23, whereas one ST218 strain (pPEKP3107) exhibited resistance to carbapenems, cephalosporins, β-lactamase/inhibitors and quinolones and harbored an ~ 59-kb IncN type MDR plasmid carrying resistance genes including blaNDM-1, dfrA14 and qnrS1. Importantly, blaNDM-1 and qnrS1 were flanked with IS26 located within the plasmid that could successfully transfer into E. coli J53. Additionally, PEKP2044 harbored an ~ 41-kb resistance plasmid located within tetA indicating resistance to doxycycline.
Conclusion
The emergence of blaNDM-1 revealed that there is great potential for ST218 Kp to become a high-risk clone for MDR-hvKp, indicating the urgent need for enhanced genomic surveillance.
Similar content being viewed by others
Introduction
Hypervirulent Klebsiella pneumoniae (hvKp), a pathotype of Klebsiella pneumoniae, was first reported in Taiwan and is a common pathogen for community-acquired pyogenic liver abscesses, distinguishing it from classical Klebsiella pneumoniae (cKp), which is usually multidrug resistant (MDR) and is highly associated with nosocomial infection [1,2,3].
Previously, hvKp was defined as hypermucoviscosity and confirmed by string test positivity [4]. Thomas and his colleagues found that aerobactin plays important roles in hypervirulence [5]. Recently, the combination of rmpA, rmpA2, iucA, iroB and peg-344 was confirmed, representing higher accuracy for defining hvKp [6]. Importantly, epidemiological investigations have demonstrated that hvKp is rapidly increasing and tends to replace cKp as the dominant pathogen in hospital- and healthcare-associated infections [12]. ST218 is a single-locus variant of ST23 that initially harbored virulence-associated genes and caused severe systemic infections but has never been systematically reported before [13].
Here, we reported for the first time that hypervirulent ST218 Kp acquired transferable blaNDM-1, which conferred MDR and hypervirulence phenotypes. Compared with ST23, we found that the genomic characteristics of ST218 are similar to those of ST23 regarding virulence and plasmid, whereas ST218 tends to acquire carbapenemases.
Methods
Enrolled ST218 Kp strains
We performed a genomic surveillance at the Peking University Third Hospital from 2017 to 2022, 1381 Klebsiella. spp were obtained, among which eleven ST218 Kp isolates (0.80%, 11/1381) were detected and further analyzed. The Vitek automated compact 2 system was used to identify the Kp isolates.
The clinical data of the infected patients was collected. Basic demographic information, specimens, infection type, antimicrobial agent exposure within 90 days, invasive incubation, metastatic infection and outcome in 30 days were collected. Additionally, the Charlson comorbidity index (CCI) and sequential organ failure assessment (SOFA) score were calculated. Community-acquired infections (CAIs) and hospital-acquired infections (HAIs) were identified as previously described [7]. Metastatic infection was defined as the presence of > 1 infection site in the same patient [14].
Antimicrobial susceptibility testing (AST)
AST was performed using the Vitek 2 system. The antimicrobial agents contained cefepime, ceftazidime, aztreonam, imipenem, meropenem, piperacillin/tazobactam, cefoperazone-sulbactam, amikacin, tobramycin, levofloxacin, ciprofloxacin, minocycline, tigecycline and trimethoprim/sulfamethoxazole. The results were interpreted according to 2021 Clinical and Laboratory Standards Institute (CLSI) guideline breakpoints, and the breakpoints for tigecycline based on the EUCAST recommendation as previously described [15]. MDR was defined as the resistance to three or more different antimicrobial categories.
String test and mucosviscosity
The hypermucoviscous phenotype was evaluated by the string test as described previously [16]. Briefly, the Kp strains were inoculated onto Columbia agar with sheep blood (PB0123A, OXOID, Bei**g, China) and cultured at 37 °C overnight. A string test was considered positive when a viscous string > 5 mm in length was generated by touching and pulling a single colony upward using a bacteriology inoculation loop.
Mucosviscosity was evaluated as previously described [17]. Briefly, Kp strains were inoculated in LB with an overnight incubation. The absorbance was measured at OD600 (preOD600). Then, 1000 μL of culture was centrifuged at 2000×g for 5 min, and the OD600 of the supernatant was evaluated (post OD600). The post/pre OD600 ratio represented the mucoviscosity.
Plasmid stability and conjugation assay
We performed a conjugation assay to evaluate the transferability of the plasmid carrying blaNDM-1. The recipient strain was classical E. coli J53, and we mixed the PEKP3107 (carrying blaNDM-1) and E. coli J5 in a 1:1 ratio and cultured in LB broth overnight [18]. Then, the MacConkey agar with a concentration of 2 mg/liter meropenem and 200 mg/liter sodium azide was prepared, on which the above mixture was further inoculated. We finally identified the transconjugants after overnight incubation. The transconjugants were evaluated by AST. Furthermore, we continuously passaged the isolates carrying blaNDM-1 and tet(A) to the 10th generation and performed AST to clarify the stability of the plasmid.
Growth curve
We selected monoclones and cultured them overnight at 37 °C. The suspension of 0.5 McF was prepared with 0.45% NaCl and then added to a 96-well plate with 3 triplicates of 10 μL per well. We further added 190 μL of LB broth per well and cultured these isolates. Finally, the absorbance at OD590 was continuously recorded by the Tecan infinite 200Pro every hour until 12 h.
Serum killing assay
The 0.5 McF bacterial solution was prepared and diluted to a final concentration of 1 × 106 CFU/ml, 25 μL of which was added to 75 μL of serum collected from healthy adults. The mixture (1 μL) was spotted on the blood plate at 0 min, 60 min, 120 min, and 180 min and cultured overnight. Viable counts were recorded. If the number of live bacteria was more than 100% after 3 hours, it was resistant, and if the opposite occurred, it was sensitive.
Biofilm formation assay
Ten microliters of 0.5 McF bacterial solution and 190 μL of LB broth were added to a 96-well plate thrice and cultured for 24 hours at 37 °C. The supernatant was absorbed and discarded, washed repeatedly with distilled water. Subsequently, 200 μL of 0.1% crystal violet dye was added, and further washed three times after 20 minutes. Finally, 200 μL of 95% ethanol was added, and the absorbance value was measured and recorded at OD 590 nm. The results were interpreted based on the absorbance value compared to the average value of the well with only LB broth (Ac) as previously described [11].
Galleria mellonella larvae lethality assay
The G. mellonella larvae lethality assay was performed as described previously [19]. Briefly, the bacterial solution of 1 × 108 CFU/ml was prepared, 10 μl of which was injected into ten larvae in each group. The infected larvae were incubated at 37 °C, and mortality was observed every 12 h for 3 days.
Whole genome sequencing (WGS) and bioinformatic analysis
First, Kp DNA was extracted for next-generation sequencing, and the preparation of libraries using Nextera technology [22]. Resistance genes, virulence genes, IS sequences and plasmid replicon types were further detected by BLAST through comparison with the ResFinder [23], Virulence Factor Database [24], IsFinder [25], and plasmidFinder [26] databases.
For phylogenetic analysis, we mapped the Illumina sequencing reads to the reference (ST23 K. pneumoniae NTUH_K2044) using Bowtie 2 v2.2.8 [27]. The single nucleotide polymorphisms (SNPs) were analyzed by Samtools v1.9. We used the iSNV calling pipeline (https://github.com/generality/iSNVcalling) to combine the SNP sites of all the ST218 Kp strains according to the reference genome (K. pneumoniae NTUH_K2044). The high-quality SNPs were collected, and the recombination sites were removed using Gubbins v2.4.1 [28]. Finally, we identified the concatenated sequences of filtered polymorphic sites that were conserved in all genomes, defined as core genome SNPs (cgSNPs), and the phylogenetic analysis was then conducted using FastTree software by the maximum likelihood method [29].
Results
Emergence of MDR hypervirulent ST218 Klebsiella pneumoniae
During the six-year surveillance, we obtained eleven ST218 Kp isolates from eleven patients out of 1381 Klebsiella isolates in total. Notably, one of the eleven Kp strains (9.1%, 1/11), PEKP3107, represented an MDR phenotype was resistant to carbapenems, cephalosporins, β-lactamase/inhibitors and quinolones (Table 1). This strain was isolated from the urine sample of a 52-year-old male patient who experienced a community-acquired infection and exposed antibiotics within 90 days. The urinary catheter and gastrostomy tube were indwelled within the patient who presented with various underlying diseases (CCI = 3) (Table 2).
In addition, another isolate named PEKP2044 presented resistance to doxycycline. This strain was isolated from sputum from a 35-year-old male patient who experienced nosocomial infection with urinary catheter intubation. In addition, none of the other nine strains presented resistance to any of the tested antimicrobial agents. All eleven isolates presented a hypermucoviscous phenotype (Table 1).
Features of resistance genes, plasmid types and virulence gene profiles
To further clarify the genomic traits of ST218 Kp, we collected a total of 29 ST218 Kp genomes, consisting of eighteen previously published and eleven we sequenced.
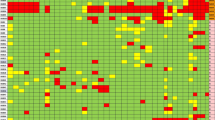
All ST218 isolates were found to possess fosA and oqxAB, which are associated with resistance to fosfomycin and quinolones. However, it should be noted that fosA and oqxAB may not necessarily confer resistance to these antibiotics in K. pneumoniae [30, 31]. Notably, the majority of the ST218 strains harbored few resistance genes. Among the 29 total ST218 Kp strains, eleven isolates (37.9%, 11/29) were predicted to be multidrug resistant. Most MDR strains harbored aac(6′)-Ib-cr, blaCTX-M-15, blaOXA-1, blaOXA-244, blaTEM-1B, qnrB1, strA, strB, sul2 and tet(A). Among our 11 isolates, PEKP3107 was predicted to be MDR and presented resistance genes including blaNDM-1 and qnrS1 and exhibited penicillin, cephalosporin, carbapenem and quinolone resistance. PEKP2044 harbored the resistance gene tet(A) and was resistant to tetracyclines (Fig. 1).
All 29 strains harbored the IncFIB type plasmid. Nine strains (31.0%, 9/29) carried the IncFII type plasmid, and eight strains (27.6%, 8/29) harbored the IncL/M plasmid replicon. The key virulence gene, iucA, was detected in 28 isolates (96.6%). A significant number of strains (92.9%, 26/29) harbored the salmochelin-associated gene (iroB) and peg-344. RmpA and rmpA2 were present in 23 and 28 isolates (79.3 and 96.6%), respectively. A majority of ST218 strains presented with the combination of iucA + iroB + peg344 + rmpA + rmpA2 (79.3%, 23/29). Only one isolate did not harbor the above five virulence genes (Fig. 1). Furthermore, we calculated that the SNP differences among these ST218 Kp isolates varied between 152 and 313, indicating that these strains were sporadic (Additional file 1: Table S1).
Coharbouring of virulence plasmid and resistance plasmid
In addition to the virulence plasmids, an ~ 59-kb MDR IncN type plasmid was also detected in the MDR isolate PEKP3107 we obtained. This plasmid, pPEKP3107–59, contained resistance genes including blaNDM-1, dfrA14 and qnrS1. This plasmid conferred to multiple resistances, including resistances to β-lactams, carbapenems, and quinolones. Moreover, blaNDM-1 and qnrS1 were located within the resistance element composed of IS26, IS3000, blaNDM-1, qnrS1 and IS26 (Figs. 2 and 3). pPEKP3107–59 showed high similarity to other blaNDM-1-carrying plasmids, such as pSCH6109-NDM (accession no. CP050859) and pJNQH116–2 (accession no. CP070900) (Fig. 3). Importantly, this IncN-type plasmid encoding blaNDM-1 could successfully transfer into E. coli J53 by conjugation, indicating the transferability of the MDR plasmid.
Comparison between plasmid pPEKP3107–59 found in this study and similar plasmids found in the online NCBI database. The outermost circle of arrows indicates the genes of the reference plasmid pPEKP3107–59 used for comparison (red: antimicrobial resistance genes; green: integrase, recombinase, and transposase genes; purple: transfer-associated genes; blue: plasmid replication and conjugation; gray: genes of other functions)
Alignments of plasmid pPEKP3107–59. The matched regions between two sequences are displayed by light blue blocks, and the identities are marked (red: antimicrobial resistance genes; green: integrase, recombinase, and transposase genes; purple: transfer-associated genes; blue: plasmid replication and conjugation; gray: genes of other functions)
Furthermore, an ~ 41-kb IncN type resistance plasmid was identified within the isolate PEKP2044. This plasmid, pPEKP2044–41 carried the resistance gene tet(A), which conferred resistance to tetracyclines; tet(A) was located within the resistance element comprised of traG, tet(A), traI and traD (Additional file 2: Figure S1). In addition, the two resistance plasmids (pPEKP3107–59 and pPEKP2044–41) remained stable after the 10th passage (Table 1).
Clinical and genomic characteristics of ST23 vs. ST218
To further explore the characteristics of ST218, a single variant of ST23, we matched ST23 as a control. The age range of the ST218-infected patients was 30 y to 82 y, while those infected with ST23 Kp ranged from 37 y to 91 y. In the ST218 group, eight patients suffered from respiratory infections (72.7%, 8/11). A significant number of the ST23 Kp strains were isolated from the blood (72.7%, 8/11), followed by sputum (18.1%, 2/11) and puncture fluid (9.1%, 1/11). Interestingly, over half of the ST218 (54.5%, 6/11) cases were defined as HAIs, whereas seven ST23 (63.6%, 7/11) cases were identified as CAIs, indicating the performance of the different infection control strategies. Six ST218 cases (54.5%, 6/11) and four ST23 cases (36.4%, 4/11) were associated with CCI ≥ 3. In addition, two ST218- and six ST23-infected patients had antimicrobial agent exposure within 90 days. Four patients (36.4%, 4/11) with ST218 Kp infection had invasive intubation, and two ST23 (18.1%, 2/11)-infected patients had a drainage tube. One patient experienced metastatic infection in both groups (Table 2).
All eleven ST218 strains in our hospital presented capsule type KL57, and all eleven ST23 isolates possessed KL1. Notably, KL57 and KL1 are closely associated with hypervirulence [1]. A majority of the ST218 isolates (90.9%, 10/11) were confirmed as O2v2 except one with O2v1, and all the ST23 isolates presented O2v2. In terms of the virulence genes, all of the ST218 and ST23 Kp strains harbored iucA, rmpA, rmpA2, iroB, peg344 and peg589, suggesting that they all belong to hypervirulent clones. Notably, all the ST23 strains but none of the ST218 strains presented colibactin-associated genes. Moreover, all the ST23 Kp strains harbored yersiniabactin-associated genes, which were present in seven ST218 Kp isolates (63.6%). For the plasmid types, all ST218 and ST23 Kp isolates harbored IncFIB-type plasmid replicons. None of the ST218 and all ST23 Kp strains harbored the IncHI1B-type plasmid replicon (Additional file 2: Figure S2, Table S2). All eleven ST23 isolates had a virulence score of 5. In contrast, seven and four ST218 strains had virulence scores of 4 and 3, respectively. ST218 and ST23 both belonged to the hypervirulent clone. Importantly, the PEKP3107 carried blaNDM-1 had a resistance score = 2, while all the other isolates showed a resistance score = 0, suggesting that ST218 tends to acquire carbapenemases (Additional file 2: Table S2).
We selected four Kp isolates, PEKP3107, PEKP2044, 219 and PEKP1095 (ST23), for further phenotype assessments. The mucoviscosity values of PEKP3107, PEKP2044, 219 and PEKP1095 (ST23 as control) were 0.18, 0.11, 0.18 and 0.27, respectively. All four Kp strains possessed strong biofilm-forming capacity and represented sensitivity to serum (Additional file 2: Table S3, Figure S3). Additionally, the growth curve showed no significant difference among the four isolates, suggesting no significant fitness cost of ST218 harboring blaNDM-1 (Additional file 2: Figure S4). Additionally, the mortality of infected G. mellonella larvae indicated no significant difference between the ST218 and ST23 Kp strains, suggesting similar hypervirulence in vivo (Additional file 2: Figure S5).
Discussion
To the best of our knowledge, this is the first report systematically demonstrating the clinical and genomic characteristics of ST218 Kp, a single-locus variant of ST23, which is commonly identified as hvKp. Notably, we reported the emergence of MDR-ST218-hvKp, which harbored key virulence-associated genes and conferred resistance to carbapenems, β-lactams, quinolones and tetracyclines due to the acquisition of a transferable MDR plasmid located with blaNDM-1, suggesting that enhanced genomic surveillance is urgently needed.
ST218 was a single-locus variant of ST23, both of which belonged to clonal Group 23 (CG23). This clone frequently presents with the pLVPK-like plasmid [32]. In this study, we found that all of the ST218 strains harbored five key virulence genes, iroB + iucA + peg344 + rmpA + rmpA2 [33]. ST218 is typically susceptible to most antibiotics, and few studies have reported its carbapenem resistance. A previous study showed that ST218 hvKp strains acquired blaOXA-48 [34]. Another study indicated that the carbapenem-resistant ST218 contributed to the acquisition of blaKPC during in vivo evolution [35]. Here, we first reported the acquisition of blaNDM-1 conferring carbapenem resistance in ST218 hvKp strains. Additionally, ST218 Kp also obtained other resistance genes, dfrA14, qnrS1 and tet(A), that formed the MDR-hvKp phenotype. Moreover, our previous study showed that the IS26 element contributed to the acquisition of resistance genes and the formation of MDR-hvKp [12, 33]. Importantly, the resistance genes blaNDM-1 and qnrS1 were flanked by IS26, indicating that enhanced genomic surveillance is essential to prevent recombination and spread. Fortunately, the difference in SNPs among ST218 strains varied more, suggesting sporadic infections within our hospital. Combined with 90 days of antibiotic exposure, the formation of MDR-ST218-hvKp might be associated with antibiotic selective pressure. It is crucial to implement active genomic surveillance to prevent MDR pathogen emergency.
Additionally, we systematically reported the clinical, microbiological and genomic features of the ST218 clone compared with those of the ST23 hypervirulent clone. ST23 is defined as the classical hypervirulent clone mainly attributed to the acquisition of the pLVPK plasmid and is closely associated with pyogenic liver abscess and bloodstream infection [36, 37]. Although most of the ST218 Kp harbored the pLVPK-like plasmid, the main infection type was respiratory infection, suggesting that clinicians should be concerned about the emergence of the ST218 hypervirulent clone. Importantly, ST218 successfully acquired transferable blaNDM-1 and became MDR-hvKp. Similarly, the convergence of MDR and hypervirulence within ST23 was also reported [33, 38], suggesting that clinicians should be aware of MDR phenotype emergence within hypervirulent clones that hinder antibacterial treatment.
In conclusion, we reported the acquisition of blaNDM-1 by typical hvKp ST218, presenting both hypervirulence and MDR phenotype, for the first time. The resistance plasmid carrying blaNDM-1 was transferable. It is of great urgency to enhance genomic surveillance to prevent their rapid propagation and evolution.
Availability of data and materials
All Klebsiella pneumoniae genome sequences in this study have been deposited in the NCBI GenBank database under the accession number PRJNA957912. The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- hvKp:
-
Hypervirulent Klebsiella pneumoniae
- cKp:
-
Classical Klebsiella pneumoniae
- MDR:
-
Multidrug resistant
- MDR-hvKp:
-
Multidrug resistant and hypervirulent Kp
- CCI:
-
Charlson comorbidity index
- SOFA:
-
Sequential organ failure assessment
- CAIs:
-
Community-acquired infections
- HAIs:
-
Hospital-acquired infections
- WGS:
-
Whole genome sequencing
- SNPs:
-
Single nucleotide polymorphisms
- cgSNPs:
-
Core genome SNPs
References
Russo TA, Marr CM. Hypervirulent Klebsiella pneumoniae. Clin Microbiol Rev. 2019;32:3.
Liu C, Shi J, Guo J. High prevalence of hypervirulent Klebsiella pneumoniae infection in the genetic background of elderly patients in two teaching hospitals in China. Infect Drug Resist. 2018;11:1031–41.
Catalán-Nájera JC, Garza-Ramos U, Barrios-Camacho H. Hypervirulence and hypermucoviscosity: Two different but complementary Klebsiella spp phenotypes? Virulence. 2017;8(7):1111–23.
Fang CT, Chuang YP, Shun CT, et al. A novel virulence gene in Klebsiella pneumoniae strains causing primary liver abscess and septic metastatic complications. J Exp Med. 2004;199(5):697–705.
Russo TA, Olson R, MacDonald U, et al. Aerobactin, but not yersiniabactin, salmochelin, or enterobactin, enables the growth/survival of hypervirulent (hypermucoviscous) Klebsiella pneumoniae ex vivo and in vivo. Infect Immun. 2015;83(8):3325–33.
Russo TA, Olson R, Fang CT, et al. Identification of biomarkers for differentiation of Hypervirulent Klebsiella pneumoniae from Classical K. Pneumoniae. J Clin Microbiol. 2018;56(9).
Liu C, Du P, **ao N, et al. Hypervirulent Klebsiella pneumoniae is emerging as an increasingly prevalent K. Pneumoniae pathotype responsible for nosocomial and healthcare-associated infections in Bei**g, China. Virulence. 2020;11(1):1215–24.
Choby JE, Howard-Anderson J, Weiss DS. Hypervirulent Klebsiella pneumoniae - clinical and molecular perspectives. J Intern Med. 2020;287(3):283–300.
Yang X, Dong N, Chan EW, et al. Carbapenem resistance-encoding and virulence-encoding conjugative plasmids in Klebsiella pneumoniae. Trends Microbiol. 2020;21
Yang P, Liu C, Wu Z, et al. Clinical outcomes, microbiological characteristics and risk factors for difficult-to-treat resistance to Klebsiella pneumoniae infection. Infect Drug Resist. 2022;15:5959–69.
Liu C, Du P, Yang P, et al. Fusion plasmid enhanced the endemic extensively drug resistant Klebsiella pneumoniae clone ST147 harbored blaOXA-48 to acquire the hypervirulence and cause fatal infection. Ann Clin Microbiol Antimicrob. 2023;22(1):11.
Liu C, Du P, Yang P, et al. Emergence of extensively drug-resistant and Hypervirulent KL2-ST65 Klebsiella pneumoniae harboring Bla (KPC-3) in Bei**g, China. Microbiol Spectr. 2022;10(6):e0304422.
Fursova NK, Astashkin EI, Ershova ON, et al. Multidrug-resistant Klebsiella pneumoniae causing severe infections in the neuro-ICU. Antibiotics (Basel). 2021;10(8)
Yang P, Wu Z, Liu C, et al. Clinical outcomes and microbiological characteristics of sequence type 11 Klebsiella pneumoniae infection. Front Med (Lausanne). 2022;9:889020.
Liu C, Du P, Yang P, et al. Emergence and inter- and Intrahost evolution of Pandrug-resistant Klebsiella pneumoniae Coharboring tmexCD1-toprJ1, blaNDM-1, and blaKPC-2. Microbiol Spectr. 2023;31:e0278622.
Shon A, Bajwa R, Russo T. Hypervirulent (hypermucoviscous) Klebsiella pneumoniae: a new and dangerous breed. Virulence. 2013;4(2):107–18.
Qian C, Zhang S, Xu M, et al. Genetic and phenotypic characterization of multidrug-resistant Klebsiella pneumoniae from liver abscess. Microbiol Spectr. 2023;4:e0224022.
Hirabayashi A, Dao TD, Takemura T, et al. A Transferable IncC-IncX3 Hybrid Plasmid Cocarrying blaNDM-4, tet(X), and tmexCD3-toprJ3 Confers Resistance to Carbapenem and Tigecycline. mSphere. 2021;6(4):e0059221.
He J, Shi Q, Chen Z, et al. Opposite evolution of pathogenicity driven by in vivo wzc and wcaJ mutations in ST11-KL64 carbapenem-resistant Klebsiella pneumoniae. Drug Resist Updat. 2023;66:100891.
Wick RR, Judd LM, Gorrie CL, et al. Unicycler: resolving bacterial genome assemblies from short and long sequencing reads. PLoS Comput Biol. 2017;13(6):e1005595.
Bankevich A, Nurk S, Antipov D, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. Journal of computational biology : a journal of computational molecular cell biology. 2012;19(5):455–77.
Lam MMC, Wick RR, Watts SC, et al. A genomic surveillance framework and genoty** tool for Klebsiella pneumoniae and its related species complex. Nat Commun. 2021;12(1):4188.
Bortolaia V, Kaas RS, Ruppe E, et al. ResFinder 4.0 for predictions of phenotypes from genotypes. J Antimicrob Chemother. 2020;75(12):3491–500.
Chen L, Yang J, Yu J, et al. VFDB: a reference database for bacterial virulence factors. Nucleic Acids Res. 2005;33(Database issue):D325–8.
Siguier P, Perochon J, Lestrade L, et al. ISfinder: the reference Centre for bacterial insertion sequences. Nucleic Acids Res. 2006;34(Database issue):D32–6.
Carattoli A, Hasman H. PlasmidFinder and in Silico pMLST: identification and ty** of plasmid replicons in whole-genome sequencing (WGS). Methods Mol Biol. 2020;2075:285–94.
Langmead B, Salzberg SL. Fast gapped-read alignment with bowtie 2. Nat Methods. 2012;9(4):357–9.
Croucher NJ, Page AJ, Connor TR, et al. Rapid phylogenetic analysis of large samples of recombinant bacterial whole genome sequences using Gubbins. Nucleic Acids Res. 2015;43(3):e15.
Price MN, Dehal PS, Arkin AP. FastTree 2--approximately maximum-likelihood trees for large alignments. PLoS One. 2010;5(3):e9490.
Bialek-Davenet S, Lavigne JP, Guyot K, et al. Differential contribution of AcrAB and OqxAB efflux pumps to multidrug resistance and virulence in Klebsiella pneumoniae. J Antimicrob Chemother. 2015;70(1):81–8.
Wang YP, Chen YH, Hung IC, et al. Transporter genes and fosA associated with Fosfomycin resistance in Carbapenem-resistant Klebsiella pneumoniae. Front Microbiol. 2022;13:816806.
Wyres KL, Wick RR, Judd LM, et al. Distinct evolutionary dynamics of horizontal gene transfer in drug resistant and virulent clones of Klebsiella pneumoniae. PLoS Genet. 2019;15(4):e1008114.
Du P, Liu C, Fan S, et al. The role of plasmid and resistance Gene Acquisition in the emergence of ST23 multi-drug resistant, Hypervirulent Klebsiella pneumoniae. Microbiology spectrum. 2022;17:e0192921.
Lev AI, Astashkin EI, Kislichkina AA, et al. Comparative analysis of Klebsiella pneumoniae strains isolated in 2012-2016 that differ by antibiotic resistance genes and virulence genes profiles. Pathog Glob Health. 2018;112(3):142–51.
Tang N, Hu J, Zhao Y, et al. In vivo evolution of carbapenem resistance in hypervirulent Klebsiella pneumoniae in a patient undergoing long-term treatment. J Antimicrob Chemother. 2022;77(2):531–3.
Gan L, Yan C, Cui J, et al. Genetic diversity and pathogenic features in Klebsiella pneumoniae isolates from patients with pyogenic liver abscess and pneumonia. Microbiol Spectr. 2022;10(2):e0264621.
Lee Y, Kim YA, Kim D, et al. Risk factors of community-onset extended-spectrum β-lactamase-producing Klebsiella pneumoniae bacteraemia in South Korea using national health insurance claims data. Int J Antimicrob Agents. 2019;54(6):723–7.
Shankar C, Jacob JJ, Vasudevan K, et al. Emergence of multidrug resistant Hypervirulent ST23 Klebsiella pneumoniae: multidrug resistant plasmid acquisition drives evolution. Front Cell Infect Microbiol. 2020;10:575289.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Key R&D Program of China (2022YFC2303201), the National Natural Science Foundation of China (82200012), the Clinical Cohort Construction Program of Peking University Third Hospital (BYSYDL2019007) and the Clinical Key Program of Peking University Third Hospital (BYSYZD2022007).
Author information
Authors and Affiliations
Contributions
ML and LC designed the study, supervised the whole project and revised the manuscript. PY performed the experiments and bioinformatic analysis, and was a major contributor in writing the manuscript. CL and PD performed the bioinformatic analysis and data collection and contributed to writing the manuscript. JY and ZW performed the experiments and statistical analysis. JZ and NS participated in the experimental performance and revised the manuscript. All authors read and approved the final manuscript.
Corresponding authors
Ethics declarations
Ethics approval and consent to participate
The protocol for this study was approved by the Peking University Third Hospital Medical Science Research Ethics Committee (M2021545). All methods have been performed in accordance with the Declaration of Helsinki. Due to the retrospective nature of the study, the need of informed consent was waived by the Peking University Third Hospital Medical Science Research Ethics Committee.
Consent for publication
Not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Additional file 1: Table S1.
The differences in SNPs of ST218 Klebsiella pneumoniae strains.
Additional file 2: Table S2.
Clinical and genomic features of ST218 vs ST23 Kp. Table S3. Microbiological phenotypes of ST218 vs ST23 Kp. Figure S1. Circular sketch map (A) and alignments (B) of plasmid ppekp2044–41. The matched regions between two sequences are displayed by light blue blocks, and the identities are marked (red: antimicrobial resistance genes; green: integrase, recombinase, and transposase genes; purple: transfer associated genes; blue: plasmid replication and conjugation; gray: genes of other functions). Figure S2. Genomic distribution of ST218 vs. ST23 Klebsiella pneumoniae strains. Red: Virulence genes; Blue: plasmid types; Dark gray: antimicrobial resistance genes. Figure S3. Serum killing of PEKP3107, PEKP2044, 219 and PEKP1095. Figure S4. Growth curve of PEKP3107, PEKP2044, 219 and PEKP1095. Figure S5. Virulence of PEKP3107, PEKP2044, 219 and PEKP1095 assessed by the Galleria mellonella infection model.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Yang, P., Liu, C., Du, P. et al. ST218 Klebsiella pneumoniae became a high-risk clone for multidrug resistance and hypervirulence. BMC Microbiol 24, 56 (2024). https://doi.org/10.1186/s12866-024-03205-8
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12866-024-03205-8