Abstract
Background
The fall webworm Hyphantria cunea is an invasive and polyphagous defoliator pest that feeds on nearly any type of deciduous tree worldwide. The silk web of H. cunea aids its aggregating behavior, provides thermal regulation and is regarded as one of causes for its rapid spread. In addition, both chemosensory and detoxification genes are vital for host adaptation in insects.
Results
Here, a high-quality genome of H. cunea was obtained. Silk-web-related genes were identified from the genome, and successful silencing of the silk protein gene HcunFib-H resulted in a significant decrease in silk web shelter production. The CAFE analysis showed that some chemosensory and detoxification gene families, such as CSPs, CCEs, GSTs and UGTs, were expanded. A transcriptome analysis using the newly sequenced H. cunea genome showed that most chemosensory genes were specifically expressed in the antennae, while most detoxification genes were highly expressed during the feeding peak. Moreover, we found that many nutrient-related genes and one detoxification gene, HcunP450 (CYP306A1), were under significant positive selection, suggesting a crucial role of these genes in host adaptation in H. cunea. At the metagenomic level, several microbial communities in H. cunea gut and their metabolic pathways might be beneficial to H. cunea for nutrient metabolism and detoxification, and might also contribute to its host adaptation.
Conclusions
These findings explain the host and environmental adaptations of H. cunea at the genetic level and provide partial evidence for the cause of its rapid invasion and potential gene targets for innovative pest management strategies.
Similar content being viewed by others
Background
The fall webworm, Hyphantria cunea Drury (Erebidae: Hyphantria), is a polyphagous pest species in forest and agricultural ecosystems; where its larvae feed on most deciduous tree leaves [1]. When trees are infested, the fall webworm consumes nearly all leaves and causes great ecological and economic impact to the forest industry [2]. H. cunea is also an invasive pest, native to North America, but has spread globally in the past seven decades [3]. Behavioural, physiological and ecological adaptations present in this species are believed to contribute to its rapid spread.
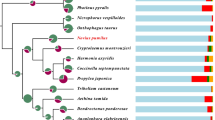
First, the fall webworm has an extremely wide range of host plants and been reported to forage on more than 600 plant species, covering nearly all types of deciduous trees, especially mulberry, boxelder, walnut, sycamore, apple, plum, cherry, and elm [4].. Insect host selection is regulated by the chemosensory systems [5], especially for polyphagous herbivores [6,7,8]. Insect chemosensory systems consist of several gene families, including odorant receptor (OR), gustatory receptor (GR), ionotropic receptor (IR), chemosensory protein (CSP) and odorant binding protein (OBP) families. These genes encode proteins that participate in host plant detection and sexual communication [9,10,11,12]. Previous investigations have suggested that the large expansions in chemosensory gene families are a possible adaptation mechanism which enables polyphagy in the lepidopteran insect Spodoptera frugiperda [13] and other taxa such as Apis mellifera, Bombyx mori and Bemisia tabaci [9, 14,15,16,17, The genome survey with k-mer analysis (Figure S1) showed that there is a small peak in depth = 22 which represented the heterozygous sequences, while the average k-mer depth was 45, and the peak indepth = 90 indicates the repetitive sequences. As a results, the tentative genome size of H. cunea was 563.96 Mb with a low heterozygosity of 0.23% and repetitive elements of 36.20% of the whole genome (Figure S1 and Table S1). The generated genome assembly of H. cunea comprises a 559.30 Mb sequence with a 3.09 Mb contig N50. It contains 198.97 Mb of repetitive elements that occupy 35.71% of the genome. After correction with RNA sequencing data from 12 samples of different tissues and stages of H. cunea, we obtained 15,319 genes using three gene prediction strategies (Figure S2), 94.42% of which could be annotated and enriched by the GO and KOG databases (Figures S3 and S4), and the distribution of Nr homologous genes with the H. cunea genome in insect species was showed in Figure S5. Moreover, 637 tRNAs, 71 rRNAs, 48 miRNAs and 300 pseudogenes were predicted from the Rfam and miRBase databases by the Infernal, tRNAscan-SE and GenBlastA software (Table S2). Further analyses showed that 94.54 and 92.96% eukaryotic conserved genes were found in the genome of H. cunea by CEGMA and BUSCO, respectively, suggesting that the genome sequence we obtained was largely complete (Tables S3-S6). The genome of H. cunea possesses a comparatively longer contig N50 among all genomes of Lepidoptera species sequenced so far, the top 4 are as follows: Operophtera brumata (6.38 Mb) [40], Spodoptera frugiperda (5.6 Mb) [48], Papilio bianor (5.5 Mb) [49], and H. cunea (3.09 Mb), further confirming the high quality of the genome sequence of H. cunea (Table 1). Homology analysis of the H. cunea genome led to the identification of 2142 pairs of one-to-one single-copy orthologs among twelve species. This ortholog dataset was used for further studies described below. Only 27 genes were specific to H. cunea, which is the smallest species-specific number among the eight lepidopteran species (Fig. 1a).
Overview of the H. cunea genome. a Types and numbers of homologous gene families among twelve species. b Maximum likelihood phylogenetic analysis among twelve insect species based on genomic data. The twelve species are Apis mellifera, Bombyx mori, Drosophila melanogaster, Helicoverpa armigera, Hyphantria cunea, Operophtera brumata, Papilio machaon, Papilio polytes, Papilio xuthus, Pieris rapae, Tribolium castaneum and Plutella xylostella. The numbers next to the nodes are the estimated node ages in million year (scale is 50.0 million years), and the colored box below indicates the geochronologic scale from Permian to Neogene RAxML was used to construct a maximum likelihood phylogenetic tree using the 2142 single-copy orthologs among twelve insects whose genome sequences were available; eight Lepidoptera were included, while Hymenoptera (A. mellifera), Coleoptera (T. castaneum) and Diptera (D. melanogaster) were used as outgroups. The results showed that all nodes were supported by strong bootstrap values of 100%, and the topology of the higher taxa was consistent with those of previous phylogenetic studies [50, 51]. The results revealed that Lepidoptera was closer to Diptera, while Hymenoptera was located at the basal branch and formed a single clade (Fig. 1b). Within Lepidoptera, Papilionoidea (butterflies) formed a single clade, and P. xylostella (Yponomeutoidea) was separated from other moth taxa (Noctuoidea, Geometroidea and Bombycoidea). H. cunea was shown to be most closely related to H. armigera, which also belongs to the superfamily Noctuoidea. These results are in agreement with those obtained from the phylogenetic studies of Lepidoptera based on morphology and molecular data [52, 53]. The phylogenetic analysis indicated that Lepidoptera diverged from Diptera approximately 244.60 million years ago, which is consistent with the previously reported divergence time [50]. In Lepidoptera, the divergent time between the moths and butterflies in our study and was at Paleogene period, which is consist with Kawahara’s work, moreover, the genetic relationship between GEOMETROIDEA and BOMBYCOIDEA were close related, and they were grouped together with NOCTUOIDEA as well [54]. H. cunea and H. armigera were estimated to have diverged at the Eocene-Oligocene boundary with a divergence time of approximately 32.07 million years ago. The period from the late Eocene to early Oligocene has been considered as an important transition time and a link between the archaic world of the tropical Eocene and the more modern ecosystems of the Miocene [55]. To further explore host adaptation, the H. cunea gene families related to chemosensory abilities (ORs, GRs, IRs, CSPs and OBPs) were studied. With the combination of de novo assembly, homology-based search and RNA sequencing annotation, 72 ORs, 46 GRs, 66 OBPs, 20 CSPs and 21 IRs were identified in the H. cunea genome (Table 2). This result increased the number of chemosensory genes in H. cunea from the previous identifications via antennal transcriptome studies, which reported 52 ORs, 9 GRs, 30 OBPs, 17 CSPs and 14 IRs [56]. For the gene families related to detoxification, 32 UGTs, 25 GSTs, 75 CCEs, 95 ABCs and 109 P450s were identified using the same strategy as above (Table 2). The numbers of chemosensory and detoxification genes in H. cunea were further compared with those of some lepidopteran insects (Table 2) [46].
Gene family expansion/contraction analyses showed that the CSP, CCE, GST and UGT gene families were expanded in H. cunea compared to the tested Lepidopteran species, as the divergence sizes were all significantly lower than the species sizes for these genes (Table 3). CSPs contribute to transportation, sensitivity and possibly the selectivity of the insect olfactory system [10]. In our study, an expansion of CSPs was detected, suggesting that they might relate to host plant selection of H. cunea, but much more testing is required. Among the detoxification gene families, UGT, CCE and GST families were found to be expanded in H. cunea (Table 3). Some studies also found that in some polyphagous species in Noctuoidea GSTs and CCEs were greatly expanded, such as H. zea, H. armigera and S. litura [45, 46].
Other major expanded gene families were hemolymph protein [57], cecropin A [58], serine protease [59], apolipophorins [60], DNA helicase [61], insulin-like growth factor [62], and yolk proteins [63, 64] (Table 3). These gene families are supported to be involved in immunity, growth and development, biomacromolecule metabolism and reproduction in insects [57,58,59, 65,66,67,68]. To further study the chemosensory and detoxification gene families that were found to be expanded, transcriptome studies on these genes were performed to explore their expression profiles in different developmental stages and tissues. The analysis of differential gene expression by pairwise comparison led to the identification of 8232 DEGs whithn the different stages RNA (eggs, second instar larvae, fourth instar larvae, pupae, and male and female adults), and 7733 DEGs within the different tissues RNA (head, thorax, leg, abdomen, antenna, and female sexual glands). Then these two DEG datasets were combined, and the duplicated sequences were removed to create a final dataset of 10,348 DEGs (Table S7). The relative expression levels of these DEGs in different tissues and stages as indicated by log10FPKM values were shown in as the Box plot in Figure S6, and the numbers of alternative splicing events was showed in Figure S7. The expression of DEG gene families (CSPs, GSTs, CCEs and UGTs) was transformed into an expression heatmap and presented in Fig. 2 to better compare their expression levels in different tissues and developmental stages. Nine of the 20 CSPs were grouped together and specifically expressed in the antennae, four CSPs were highly expressed in pupae relative to other stages, while two CSPs were specifically expressed in the sex gland (Fig. 2a). In the expanded detoxification gene families CCE, GST and UGT, some genes were highly expressed in the fourth larval instar (Fig. 2b, c and d), which is the peak period of H. cunea foraging behavior [1].
Expression heatmap of the expanded gene families in different tissues and stages. The colors represent the level of gene expression from low (purple) to high (yellow) as shown by log10 (FPKM + 0.000001). E (eggs), L2 (second-instar larvae), L4 (fourth-instar larvae), P (pupae), F (female adults), M (male adults); An (antenna), H (head), T (thorax), Ab (abdomen), L (leg), and Sg (female sexual gland). a Expression heatmap of the CSP gene family. The transcripts that were grouped together and specifically expressed in the antennae are EVM0003477, EVM0000271, EVM0002854, EVM0002317, EVM0009785, EVM0004106, EVM0004147, EVM0005331 and EVM0008758. The transcripts that were highly expressed in pupae are EVM0014103, EVM0014851, EVM0003329 and EVM0014642. The transcripts that were specifically expressed in the sex gland are EVM0010490 and EVM0014431. b Expression heatmap of the CCE gene family. c Expression heatmap of the GST gene family. d Expression heatmap of the UGT gene family. The heatmaps were constructed with HemI (windows_1_0_win32_x64) using the DEGs of each expanding family. The expression levels are presented by the color bar from low expression as green with negative numbers and high expression level as yellow with positive numbers Next, a positive selection analysis based on the homolog genes was performed on the genome of H. cunea to gain a better understanding of the mechanisms in its host selection. The branch-site model showed that 39 genes were under significant positive selection pressure (LRT, p < 0.05), of which 13 were nutrient regulation genes reported to be involved in the metabolism of lipids, carbohydrates, vitamins and amino acids (Table 4). Many studies have shown that nutrient regulation in herbivorous insects is shaped by natural selection [69, 70]. Significant positive selection pressure was also detected in HcunP450 (EVM0009687), a member of the major detoxification-related gene family P450 (Table 4), consistent with a previous study reported that P450s could mediate insect resistance to many classes of insecticides [71]. HcunP450 was most similar to the cytochrome P450 CYP306A1 of the cotton bollworm H. armigera (AID54855.1), with 81.63% identity at the amino acid level. The expression of HarmP450 CYP306A1 was found to be induced by 2-tridecanone, and to mediate cotton bollworm development [72]. The CYP306A1 gene family was also shown to play an essential role in ecdysteroid biosynthesis during insect development [73], in fluoride resistance of B. mori [74]. Thus, the positive selection of HcunP450 CYP306A1 might reflect the rapid development of insecticide resistance in H. cunea. However, it is needed to determine whether it is caused by long-term host adaptation or by rapid evolution due to the extensive use of insecticides in recent years.
Our gut microbiota sequencing of H. cunea yielded 8.65 GB of valid data after filtering of H. cunea genome sequences and produced 28,846,959 clean reads and 151,448 contigs with a total length of 520.68 Mb after de novo assembly (Table S8). Based on the alignment of sequencing reads to the NCBI RefSeq database, the microorganism composition was annotated (Table S9) and analyzed, and the microbes were grouped into taxonomic categories from kingdom to species level. We found 324 kingdoms, 135 phyla, 13 classes, 244 orders, 157 families, 200 genera, and 78 species in the larval gut of H. cunea. At the phylum level, the H. cunea gut microbiota was dominated by Proteobacteria (71.33% of the total midgut bacteria contigs), followed by Euryarchaeota and Firmicutes (8.40 and 6.10% of the contigs, respectively) and to a lesser extent, Tenericutes, Actinobacteria, Cyanobacteria and Bacteroidetes; other phyla were less than 1% of the total contigs (Fig. 3a). At the class level, Gammaproteobacteria, Betaproteobacteria, Halobacteria and Clostridia comprised 77% of the contigs (Fig. 3b), while Enterobacteriales, Halobacteriales and Burkholderiales comprised 60% of all contigs at the order level (Fig. 3c). The three most abundant families were Enterobacteriaceae, Halobacteriaceae and Burkholderiaceae (50.86, 6.16, and 4.58% of total contigs, respectively) (Fig. 3d). At the genus level, microorganisms were rich in Klebsiella, Halovivax and Burkholderia (37.92, 4.75 and 4.32% of total contigs, respectively) (Fig. 3e). Klebsiella oxytoca was the most abundant species in the midgut of H. cunea, followed by Halovivax ruber, Mannheimia haemolytica, and Burkholderia vietnamiensis (Fig. 3f).
Taxonomic-category-based microbiota composition in the H. cunea larval midgut. a Microbiota proportion at the phylum level. b Microbiota proportion at the order level. c Microbiota proportion at the class level. d Microbiota proportion at the family level. e Microbiota proportion at the genus level. f Microbiota proportion at the species level Our metagenomic analysis led to the identification of 102,787 nonredundant protein-coding genes with an average length of 300 bp (30.80 Mb total length) in the microbiota of the H. cunea larval gut. Gene functional annotation based on KEGG pathways showed that the most abundant function in the metagenome was metabolic function, representing 45.16% of all KEGG functions in the H. cunea gut microbiota. KEGG iPath 2 analysis showed that the metabolic activities of the H. cunea gut bacteria were associated with digestion, nutrition and detoxification, including metabolism of energy, carbohydrates, amino acids, lipids, cofactors, vitamins, glycans, xenobiotics, and terpenoids. The most enriched functions within these activities were “Folding, sorting and degradation”, representing 15.35% of all KEGG pathways, followed by “Signal transduction” (11.08%). The nutrient metabolism functions that could be provided by gut microbiota were “Carbohydrate metabolism” (8.83%), “Amino acid metabolism” (7.09%), “Energy metabolism” (6.59%), “Nucleotide metabolism” (4.55%), “Lipid metabolism” (3.90%), “Glycan biosynthesis and metabolism” (1.93%) and “Metabolism of cofactors and vitamins” (2.72%). In addition, genes in the gut microbiota were found with functions related to “Xenobiotics biodegradation and terpenoid metabolism” (2.09%) and “Biosynthesis of other secondary metabolites” (0.31%) (Figs. 4 and S9 and Table S10).
A total of 336 enzymes associated with cellulose and hemicellulose hydrolysis were identified in the intestinal flora of H. cunea based on the Carbohydrate-Active EnZyme (CAZy) database, including 42 auxiliary activities (AAs), 68 carbohydrate binding modules (CBMs), 75 carbohydrate esterases (CEs), 68 glycoside hydrolases (GHs), 82 glycosyltransferases (GTs) and one polysaccharide lyase (PL) (Figure S8 and Table S11). The results indicate that the gut microbes of H. cunea were most likely involved in cellulose degradation. By sequence alignment, we also predicted 55, 256 and 236 genes possibly encoding for glutathione S-transferases, esterases, and P450s, respectively (Table S12). Notably, one gene family related to silk production, Kazal-type serine proteinase inhibitors (KSPIs) [75], showed an expansion among the tested orthologous gene groups, which implies that silk-related genes might also have a role in the environmental adaptation to larval development of H. cunea. Hence, we performed further studies on silk-web-related genes. The silk gland is a long paired organ of the fall webworm. It specializes in the synthesis and secretion of silk proteins (Fig. 5a) and quickly atrophies after the onset of adulthood. The anatomy of the silk gland in the fall webworm is quite similar to that of B. mori, and consists of three functionally distinct regions: the anterior silk gland (ASG), middle silk gland (MSG) and posterior silk gland (PSG) [76]. Thirty-three silk-gland-related genes were identified in H. cunea (Table 5) through a homologous search against those from other Lepidopteran silk glands in previous studies [76,77,78], including 3 silk protein genes, 4 silk regulation genes and 26 protease inhibitor genes. In B. mori, the silk protein is composed of a ((Fib-H) - (Fib-L))6 -P25 fibroin complex and held together by the protein sericin [79]. Here, three fibroin structure genes, HcunFib-H, HcunFib-L and HcunP25, were identified from the H. cunea genome, but our results showed that no sericin genes were annotated in the H. cunea genome; however, some silk regulation genes, such as silk gland factors (SGFs), fibroin-modulator-binding protein-1 (FMBP-1) and fibroinase, were identified in the H. cunea genome. Moreover, several protease inhibitors, such as kazal-type serine protease inhibitors, pacifastin-related serine protease inhibitor (pacifastin), phosphatidylethanolamine-binding protein, alpha-2-macroglobulin (A2M), cysteine proteinase inhibitor, carboxypeptidase inhibitor, cystatin, serpins and proteasome inhibitor genes, were identified.
Silencing results of HcunFib-H and phenotype analysis. a Anatomy of the larval silk gland of H. cunea. ASG indicates anterior silk gland, MSG indicates middle silk gland and PSG indicates posterior silk gland. b RT-qPCR results 4 days after dsRNA injection for RNAi. Statistical differences were evaluated by t-tests. *** p < 0.001, n.s., not significant. Data indicate the means + SEM, N = 5. The expression levels were normalised by the expression of β-actin gene in different treatment samples. c & d Diameter of the silk ball after RNAi; statistical differences were evaluated by t-tests. *** p < 0.001, n.s., not significant, scale bar = 0.5 mm. e & f Comparison of silk spinning between wild-type and dsHcunFib-H-injected insects Because silk is a structural material and plays a crucial role in the survival of many insects, the extraordinary mechanical properties of silk are often explained in adaptive terms [80]. Fibroin is the key component of silk; it determines both the quantity and the structure of a silk web [81]. Here, three structural protein genes, HcunFib-H, HcunFib-L and HcunP25, were chosen for RNAi experiments to explore the mechanism of web production in H. cunea because of their involvement in silk production. We targeted three fibroin genes for silencing and measured their expression levels by qRT-PCR 4 days after injection (Fig. 5b). In comparison with the noninjected groups, there were no significant changes in the expression of GFP, HcunFib-L and HcunP25 (p > 0.05), while the relative expression of HcunFib-H was dramatically decreased (p < 0.001). The different expression levels among the three genes might be one of reasons that resulted in the differences in RNAi as the expression of HcunFib-H was much higher than those of HcunFib-L and HcunP25. Within 10 days after the injection, the average diameters of the silk balls in the different treatments were as follows: that of the noninjected wild type was 3.45 ± 0.12 mm; dsGFP-injected, 3.39 ± 0.14 mm (p = 0.64 > 0.05); dsHcunFib-L-injected, 3.41 ± 0.15 mm (p = 0.79 > 0.05); dsHcunP25-injected, 3.25 ± 0.14 mm (p = 0.18 > 0.05) and dsHcunFib-H-injected, 1.67 ± 0.09 mm (p < 0.0001) (Fig. 5c and d). There was a significant decrease in the quantity of silk in the dsHcunFib-H injected group, which was consistent with the dramatic decrease in the gene expression of HcunFib-H of the dsHcunFib-H-injected group after RNAi. The silencing of the silk structure protein gene Fib-H led to less silk production and damaged the leaf-silk shelter structure of the fall webworm by breaking the silk-leaf connections (Fig. 5e and f), suggesting that HcunFib-H contributes significantly to the formation of fibroin, to related web-producing behaviors and to the silk-web-related adaptations of H. cunea. In this study, the genome of the fall webworm we obtained was of high integrity by PacBio sequencing. And compared with other publicly available Lepidopteran genomes (Plutella. xylostella, Papilio polytes, Papilio machaon, Papilio xuthus, Pieris rapae, H. armigera, O. brumata, B. mori, S. frugiperda and P. bianor), the H. cunea genome possesses a comparatively longer contig N50 (only smaller than the genome of O. brumata, S. frugiperda and P. bianor). The large genome size of O. brumata could be explained to a large extent by its higher repeat content, containing 53.5% repetitive elements in O. brumata genome (35.7% in H. cunea and 38.4% in B. mori genomes) [40]. However, the large genome of H. cunea is more likely to be caused by a larger average intron size, the mechanism is worthy of further study, because the average intron size of the H. cunea genome was 1491 bp, much larger than 1082 bp of B. mori and 139 bp of O. brumata. A similar phenomenon was also reported in the Locusta migratoria genome [82]. According to the result of the phylogeny of Lepidoptera, H. cunea and H. armigera were estimated to have diverged at the Eocene-Oligocene boundary, while from the late Eocene to early Oligocene, with the end of a continuous cooling event [83], deciduous trees that were better able to cope with large temperature changes began to overtake evergreen tropical species [84]. In North America, where H. cunea is native, litchi and cashew nut were the dominant trees in the early Oligocene [85] . With the expansion of temperate deciduous forests during this epoch, the food sources of the fall webworm increased, which might have contributed to the expansion of the host range of H. cunea. The CSP, CCE, GST and UGT gene families were expanded in H. cunea compared to the tested Lepidopteran species, similar expansions of the chemosensory gene family have also been detected in other insect genomes [13, 154] and Benchmarking Universal Single-Copy Orthologs (BUSCO v2.0) [155], were used to assess the completeness of the WTDBG assembly. The specific repetitive sequence database was used to predict repeat sequences. A de novo repeat library of H. cunea was constructed by LTR_FINDER v1.05 [156], MITE-Hunter [157], RepeatScout v1.0.5 [158] and PILER-DF v2.4 [159]; then, it was classified by PASTE Classifier [160] and combined with the Repbase transposable element library to act as the final library. Afterward, RepeatMasker v4.0.6 [161] was used to find the homologous repeats in the final library. tRNAscan-SE v1.3.1 [162] was used to search for tRNA coding sequences. rRNA and microRNA were identified by Infernal v1.1 [163] based on the Rfam database and miRBase database. The pseudogene were predicted by two steps: Firstly, GenBlastA v1.0.4 [164] was applied to identify the candidate pseudogene by homologous searching against genome data. Secondly, GeneWise v2.4.1 [165] was performed to search for immature termination and frameshift mutation of pseudogene . To identify protein-coding sequences, a combination of ab initio gene prediction, homology-based prediction and unigene-based methods were used as annotation pipelines. Genscan [166], Augustus v2.4 [167], GlimmerHMM v3.0.4 [168], GeneID v1.4 [169] and SNAP [170] were used to predict the protein-coding sequences. Four species (Amyelois transitella, Bombyx mori, Helicoverpa armigera and Plutella xylostella) were used to complete homology-based gene prediction with GeMoMa v1.3.1 [171]. Reference transcriptome assembly was performed by HISAT v2.0.4 and StringTie [172], and gene prediction was performed by TransDecoder v2.0 [173] and GeneMarkS-T v5.1 [174]. The de novo transcriptome was completed by PASA v2.0.2 [175]. Finally, all the results from three gene prediction methods (GeMoMa, TransDecoder v2.0 and GeneMarkS-T) were integrated by EVidenceModeler (EVM) v1.1.1 [176] and annotated by PASA v2.0.2. Gene functions were assigned according to the best-match BLASTp alignments in the NR databases, KOG, TrEMBL and Kyoto Encyclopedia of Genes and Genomes (KEGG). GO annotations were obtained by Blast2GO based on the results of alignment to NR. Moreover, we also performed enrichment analyses of the Clusters of Orthologous Groups of proteins (COG), GO terms and KEGG pathways. The most updated genome sequences of twelve sequenced insects (Apis mellifera, Bombyx mori, Drosophila melanogaster, Helicoverpa armigera, Papilio machaon, Papilio polytes, Papilio xuthus, Pieris rapae, Plutella xylostella, Tribolium castaneum, Hyphantria cunea, and Operophtera brumata) were used to infer gene orthology and construct the phylogenetic tree, the details of these genomic datasets we used in this study were showe in Table S13. After downloading the annotated coding sequences from NCBI, the longest protein sequences per gene were extracted to perform a best reciprocal hit (BRH) analysis by all-v-all BLAST using an E-value equal to 1E− 05 to identify orthologous genes among the twelve species by OrthoMCL 5 [177]. The longest open reading frames (ORFs) for the longest transcript pairs across the twelve species were extracted by a Perl script, and tORFs in each orthologous set were aligned using PRANK [178] with the following parameters: −f = fasta -F -codon -noxml -notree -nopost. The alignment for each locus was trimmed by Gblocks v 0.91b [179] (Parameters: −t = c − b3 = 1 − b4 = 6 − b5 = n) to reduce the rate of false positive predictions by filtering out sequencing errors, incorrect alignments and no-orthologous regions based on codons [180]. After trimming, alignments of less than 120 bp were removed. The single-copy orthologous genes were concatenated into one supergene, and the best amino acid substitution model was estimated. RAxML v. 8.0.26 [181] was used to construct the phylogenetic tree based on the supergene under the LG + I + G + F model with 1000 bootstrap replicates. The divergence times among species were estimated by R8s v. 1.7.1 [182] with a node dating approach that used three fossil records as the most recent common ancestor. The three fossil records we used in this study were the oldest definitive beetle (Coleopsis archaica gen. et sp. Nov., 298.9 to 295.0 Ma), the oldest fossil Diptera (such as: Anisinodus crinitus n. gen., n. sp., 247.2 to 242.0 Ma) and the oldest fossil Rhopalocera (Praepapilio Colorado n. g., n. sp., P. gracilis n. sp., and Praepapilioninae. Riodinella nympha n. g., n. sp., 46.2 to 40.4 Ma), respectively [183,184,185]. CAFE [186] was used to examine the expansion and contraction of gene families among the twelve species. The results of the orthologous gene identification were filtered by CAFE’s built-in script, and the global parameter λ was estimated by the maximum likelihood method. Comparing divergence size and species size calculated by CAFE could determine whether expansion had occurred. The divergence size indicates the ancestral gene family size for each node in the phylogenetic tree, and the species size indicates the gene number in the homologous gene family. When the divergence size is smaller than the species size, the gene family is expanding. Additionally, for each gene family, a conditional P-value was calculated, and gene families with P-values < 0.05 were considered to have significantly expanded or contracted. A branch-site model (parameters: Null hypothesis: model = 2, NSsites = 2, fix_omega = 1, omega = 1; alternative hypothesis: model = 2, NSsites = 2, fix_omega = 0, omega = 1) in PAML [187] was used to identify the genes with positively selected sites in the fall webworm genome using our tree topology as the guide tree. Then, likelihood ratio tests (LRTs) were performed to detect positive selection on the foreground branch. Only those genes with LRT P-values less than 0.05 were inferred as positively selected. RNA sequencing was performed on different developmental stages and tissues of H. cunea. The following developmental stages were selected for the transcriptome analyses: eggs, second instar larvae, fourth instar larvae, pupae, and male and female adults. The following tissues were used for the tissue transcriptome experiment: head, thorax, leg, abdomen, antenna, and female sexual glands. For each group, fifteen individuals were mixed for RNA extraction, and three biological replicates were produced for each sample. Total RNA was isolated from the homogenized samples using TRIzol reagent (Invitrogen, Carlsbad, CA, USA) according to the manufacturer’s protocols. After extraction, total RNA was assessed with the NanoDrop 2000 (Thermo Fisher Scientific, Waltham, MA, USA) and the Agilent Bioanalyzer 2100 System (Agilent Technologies, CA, USA) to verify the integrity and quality of RNA. After each sample was quantified, the libraries were built and sequenced on the Illumina HiSeq X Ten platform. After filtering, clean reads were mapped to the reference genome sequence obtained in this study with Hisat2 tools [188]. Only reads with a perfect match or one mismatch were retained for further analysis. Cufflinks counts the expression of each gene and reports it in fragments per kilobase of transcript per million fragments mapped (FPKM) [189]. For each sequenced library, the read counts were adjusted by the edgeR package [190] with one scaling normalized factor. Differentially expressed gene (DEG) analysis within two sample groups (stages and tissues) was performed using the EBSeq R package [191], and then the false discovery rate (FDR) was performed based on the Benjamini-Hochberg (BH) procedure [192] to correct the P value of the identified datasets, with the standard of FDR ≤ 0.01 and fold change (FC) ≥ 2 to remove the false positive datasets. To test whether symbiotic microbes facilitate environmental adaptation in H. cunea, detailed profiles of the gut microflora were obtained by metagenomic sequencing. Midgut samples were collected from ten last-instar larvae of H. cunea from the same wild population on their host plant (Quercus mongolica) and preserved in RNAlater. DNA was extracted from a mixture of ten gut samples using an effective gut microbiota DNA extraction kit (QIAamp DNA Stool Mini Kit; Qiagen) and stored at − 20 °C. A paired-end gut microbiota DNA library was built using the NEBNext DNA Library Prep Mast Mix Set for Illumina (New England Biolabs, Ipswich, MA, USA). Sequencing was then performed on the Illumina HiSeq platform. The raw reads were checked and filtered by the following methods: 1) reads with adapters were removed; 2) reads with low-quality and N bases (quality value ≤10) were removed; 3) to gain a clearer understanding of the bacterial genome data, the host genome data were filtered out by eliminating fall webworm genome sequences. Kraken [193] was used for the taxonomic identification and relative abundance calculations, and the NCBI Reference Sequence Database (RefSeq), which includes high-quality bacterial, archaea and virus data, could further filter the nonbacterial genome sequences. The microbiota composition was visualized by Krona [194] and Python scripts. De novo assembly was performed by IDBA-UD [195] (parameter: --mink:21, −-maxk:101, −-step:20, −-pre_correction), resulting in sequences greater than 500 bp. The assembly quality was assessed by QUAST [196]. MetaGeneMark [197] was used to perform ab initio gene prediction with the default settings. Prophage prediction was performed by BLAST (E-value: 1E-05) with a local database based on the ACLAME database. Transposable elements, including DNA transposons, long terminal repeats (LTRs), long interspersed elements (LINEs) and short interspersed elements (SINEs), were identified by using RepeatMasker v 4.0.5 and RepeatProteinMasker [161]. A nonredundant data set was outputted by CD-HIT [198] with a minimum coverage cut-off of 0.9 for the shorter sequences. All genes in our nonredundant dataset were translated into amino acid sequences and aligned to relevant databases: NR, COG, KEGG, Swiss-Prot, CAZy and ARDB by BLASTP (E-value ≤1E-05). Blast2GO was used to obtain GO annotations, and HMMER v 3.0 [199] was used to annotate sequences in our dataset from the Pfam database. To study the web-producing mechanism in the fall webworm, silk-gland-related genes were identified by analyzing the H. cunea genome and the silk gland transcriptome produced in this study. Three genes encoding structural proteins, fibroin heavy (Fib-H), fibroin light (Fib-L) and protein 25 (P25), were silenced by RNA interference (RNAi) to examine their biological functions. RNAi was performed by injecting the corresponding gene-specific double-stranded RNAs (dsRNAs), and green fluorescent protein (GFP) was used as a negative control. The dsRNAs for HcunFib-H, HcunFib-L, HcunP25 and GFP were synthesized by using the MEGAscript RNAi Kit (Ambion, Austin, TX, USA) following the manufacturer’s procedure and purified by lithium chloride precipitation. After quantification with a NanoDrop 2000 (Thermo Fisher Scientific, Wilmington DE, USA) and 1% agarose gel electrophoresis, the dsRNA of the four genes was stored at − 80 °C before use. Then, newly molted third-instar larvae were injected with 4 μg of targeted dsRNA in 1 μL into the abdomen using a Nanoliter 2000 injector (World Precision Instruments, Sarasota, FL, USA). In total, 20 individuals were injected, divided into four plastic boxes (20 cm × 10 cm × 5 cm) and fed fresh mulberry leaves (6 g per box per day); of these, 15 individuals were used to observe the phenotype (N = 3), while 5 individuals were used for RT-qPCR validation (N = 5). The effect of RNAi was examined by RT-qPCR 4 days after injection; each cDNA sample was quantified based on the total RNA (2 μg) from the 5 insects separately before reverse transcription (SuperScript™ III First-Strand Synthesis SuperMix), and β-actin was employed as an internal control. RT-qPCR was performed on a StepOnePlus Real-Time PCR Detection System (Bio-Rad, Hercules, CA, USA) using TransStar Tip Top Green qPCR Supermix (TransGen Biotech, Bei**g, China). The silk web was collected from each box within 10 days after injection. Because the silk filaments were difficult to quantify, they were rolled into a tight ball, and the diameter of the silk ball was used to calculate the silk quantity. The RT-qPCR data were analyzed by the 2-ΔΔCT method. The primers used in this study are listed in Table S14, and the efficiency of each primer pair was tested before the RT-qPCR experiments.Results
Overview of genome assembly and annotation
Phylogeny of Lepidoptera
Expansion of chemosensory and detoxification gene families
DEG analysis in different stages and tissues
Positive selection on genes related to nutrient metabolism and detoxification
Compositional diversity of the gut microbiota
Functional annotation of the leaf-eating caterpillar gut metagenome
Silk-web-related genes
Silencing of silk fibroin genes and phenotype analysis
Discussion
Repeats and noncoding RNAs
Gene prediction and functional annotation
Orthologous gene families
Phylogenetic tree and divergence times
Gene family expansion/contraction
Positive selection analyses
Transcriptome analysis of different stages and tissues
Metagenomic sequencing and analysis
RNA interference with silk fibroin genes
Availability of data and materials
The genome data of H. cunea have been deposited in the SRA under the accession number SUB5033887.
Abbreviations
- A2M:
-
Alpha-2-macroglobulin
- AAs:
-
Auxiliary activities
- ABCs:
-
ATP-binding cassette transporters
- ASG:
-
Anterior silk gland
- CAZy:
-
Carbohydrate-Active enZYmes
- CBMs:
-
Carbohydrate binding modules
- CCEs:
-
Carboxyl/choline esterases
- CEs:
-
Carbohydrate esterases
- COG:
-
Cluster of Orthologous Groups
- CSPs:
-
Chemosensory Proteins
- DEGs:
-
Differential expression genes
- dsRNAs:
-
double-stranded RNAs
- Fib-H:
-
Fibroin heavy chain gene
- Fib-L:
-
Fibroin light chain gene
- FMBP-1:
-
Fibroin-modulator-binding protein-1
- FPKM:
-
Kilobase of transcript per million fragments mapped
- GFP:
-
Green fluorescent protein
- GHs:
-
Glycoside hydrolases
- GRs:
-
Gustatory Receptors
- GSTs:
-
Glutathione S-transferases
- GTs:
-
Glycosyltransferases
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- KOG:
-
euKaryotic Orthologous Groups
- KSPIs:
-
Kazal-type serine proteinase inhibitor
- LINE:
-
Long Interspersed Elements
- LRTs:
-
Likelihood ratio tests
- LTR:
-
Long Terminal Repeat
- MSG:
-
Middle silk gland
- NR:
-
NCBI non-redundant
- OBPs:
-
Odorant Binding Proteins
- ORs:
-
Odorant Receptors
- P25:
-
Protein 25
- Pacifastin:
-
Pacifastin-related serine protease inhibitor
- PBP:
-
Phosphatidylethanolamine-binding protein
- Pfam:
-
Protein family
- PLs:
-
Polysaccharide lyases
- PSG:
-
Posterior silk gland
- RefSeq:
-
Reference Sequence Database
- SGFs:
-
Silk gland factors
- SINE:
-
Short Interspersed Elements
- SPI 2:
-
Silk proteinase inhibitor 2
- UGTs:
-
UDP glycosyltransferases
References
Schowalter T, Ring D. Biology and Management of the Fall Webworm, Hyphantria cunea (Lepidoptera: Erebidae). J Integr Pest Manage. 2017;8(1):7.
Ge X, He S, Zhu C, Wang T, Xu Z, Zong S. Projecting the current and future potential global distribution of Hyphantria cunea (Lepidoptera: Arctiidae) using CLIMEX[J]. Pest Manag Sci. 2019;75(1):160-69.
Cocquempot C, Lindelöw A. BIORISK-biodiversity and ecosystem risk assessment, vol. 4. Sofia: Pensoft Publishers; 2010. p. 193–218.
Sullivan GT, Karaca I, Ozman-Sullivan SK, Kara K. Tachinid (Diptera: Tachinidae) parasitoids of overwintered Hyphantria cunea (Drury)(Lepidoptera: Arctiidae) pupae in hazelnut plantations in Samsun province, Turkey. J Ent Res Soc. 2012;14:21–30.
Chapman R. Chemosensory regulation of feeding. Regulatory mechanisms in insect feeding: Springer; 1995. p. 101–36.
Qin J, Wang C. The relation of interaction between insects and plants to evolution[J]. Acta Ecol Sin. 2001;44(3):360-65.
Ishikawa S, Hirao T, Arai N. Chemosensory basis of hostplant selection in the silkworm. Entomologia Experimentalis et Applicata. 1969;12(5):544–54.
CORBET SA. Insect chemosensory responses: a chemical legacy hypothesis. Ecol Entomol. 1985;10(2):143–53.
Sánchez-Gracia A, Vieira F, Rozas J. Molecular evolution of the major chemosensory gene families in insects. Heredity. 2009;103(3):208.
Leal WS. Odorant reception in insects: roles of receptors, binding proteins, and degrading enzymes. Annu Rev Entomol. 2013;58(1):373–91.
Benton R. Multigene family evolution: perspectives from insect chemoreceptors. Trends Ecol Evol. 2015;30(10):590–600.
Simon J-C, d’Alencon E, Guy E, Jacquin-Joly E, Jaquiery J, Nouhaud P, et al. Genomics of adaptation to host-plants in herbivorous insects. Brief Function Genomics. 2015;14(6):413–23.
Gouin A, Bretaudeau A, Nam K, Gimenez S, Aury JM, Duvic B, et al. Two genomes of highly polyphagous lepidopteran pests (Spodoptera frugiperda, Noctuidae) with different host-plant ranges. Sci Rep. 2017;7(1):11816.
Robertson HM, Wanner KW. The chemoreceptor superfamily in the honey bee, Apis mellifera: expansion of the odorant, but not gustatory, receptor family. Genome Res. 2006;16(11):1395–403.
Wanner K, Robertson H. The gustatory receptor family in the silkworm moth Bombyx mori is characterized by a large expansion of a single lineage of putative bitter receptors. Insect Mol Biol. 2008;17(6):621–9.
Obiero GF, Mireji PO, Nyanjom SR, Christoffels A, Robertson HM, Masiga DK. Odorant and gustatory receptors in the tsetse fly Glossina morsitans morsitans. PLoS Negl Trop Dis. 2014;8(4):e2663.
Opachaloemphan C, Yan H, Leibholz A, Desplan C, Reinberg D. Recent advances in behavioral (Epi) genetics in Eusocial insects. Annu Rev Genet. 2018;52:489–510.
Zeng Y, Yang YT, Wu QJ, Wang SL, **e W, Zhang YJ. Genome-wide analysis of odorant-binding proteins and chemosensory proteins in the sweet potato whitefly, Bemisia tabaci. Insect Sci. 2019;26(4):620–34.
Despres L, David J-P, Gallet C. The evolutionary ecology of insect resistance to plant chemicals. Trends Ecol Evol. 2007;22(6):298–307.
Edger PP, Heidel-Fischer HM, Bekaert M, Rota J, Glöckner G, Platts AE, et al. The butterfly plant arms-race escalated by gene and genome duplications. Proc Natl Acad Sci. 2015;112(27):8362–6.
Rane RV, Walsh TK, Pearce SL, Jermiin LS, Gordon KH, Richards S, et al. Are feeding preferences and insecticide resistance associated with the size of detoxifying enzyme families in insect herbivores? Curr Opin Insect Sci. 2016;13:70–6.
Hidaka T. Adaptation and speciation in the fall webworm; 1977.
Rajagopal R. Beneficial interactions between insects and gut bacteria. Indian J Microbiol. 2009;49(2):114–9.
Engel P, Moran NA. The gut microbiota of insects–diversity in structure and function. FEMS Microbiol Rev. 2013;37(5):699–735.
Krishnan M, Bharathiraja C, Pandiarajan J, Prasanna VA, Rajendhran J, Gunasekaran P. Insect gut microbiome–An unexploited reserve for biotechnological application. Asian Pac J Trop Biomed. 2014;4:S16–21.
Dillon R, Dillon V. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49(1):71–92.
Dillon R, Charnley K. Mutualism between the desert locust Schistocerca gregaria and its gut microbiota. Res Microbiol. 2002;153(8):503–9.
Wernegreen JJ. Mutualism meltdown in insects: bacteria constrain thermal adaptation. Curr Opin Microbiol. 2012;15(3):255–62.
Mueller UG, Mikheyev AS, Hong E, Sen R, Warren DL, Solomon SE, et al. Evolution of cold-tolerant fungal symbionts permits winter fungiculture by leafcutter ants at the northern frontier of a tropical ant–fungus symbiosis. Proc Natl Acad Sci. 2011;108(10):4053–6.
Montllor CB, Maxmen A, Purcell AH. Facultative bacterial endosymbionts benefit pea aphids Acyrthosiphon pisum under heat stress. Ecol Entomol. 2002;27(2):189–95.
Russell JA, Moran NA. Costs and benefits of symbiont infection in aphids: variation among symbionts and across temperatures. Proc R Soc B Biol Sci. 2005;273(1586):603–10.
Perlman SJ, Kelly SE, Hunter MS. Population biology of cytoplasmic incompatibility: maintenance and spread of Cardinium symbionts in a parasitic wasp. Genetics. 2008;178(2):1003–11.
Loewy KJ, Flansburg AL, Grenis K, Kjeldgaard MK, Mccarty J, Montesano L, et al. Life history traits and rearing techniques for fall webworms (Hyphantria cunea Drury) in Colorado. J Lepidopterists' Soc. 2013;67(3):196–205.
Rehnberg BG. Heat retention by webs of the fall webworm Hyphantria cunea (Lepidoptera: Arctiidae): infrared warming and forced convective cooling. J Therm Biol. 2002;27(6):525–30.
Mondal M. The silk proteins, sericin and fibroin in silkworm, Bombyx mori Linn.,-a review. Caspian J Environ Sci. 2007;5(2):63–76.
Devi R, Deori M, Devi D. Evaluation of antioxidant activities of silk protein sericin secreted by silkworm Antheraea assamensis (Lepidoptera: Saturniidae). J Pharm Res. 2011;4(12):4688–91.
Zurovec M, Kludkiewicz B, Fedic R, Sulitkova J, Mach V, Kucerova L, et al. Functional conservation and structural diversification of silk sericins in two moth species. Biomacromolecules. 2013;14(6):1859–66.
Stewart RJ, Wang CS. Adaptation of caddisfly larval silks to aquatic habitats by phosphorylation of H-fibroin serines. Biomacromolecules. 2010;11(4):969–74.
**a Q, Zhou Z, Lu C, Cheng D, Dai F, Li B, et al. A draft sequence for the genome of the domesticated silkworm (Bombyx mori). Science. 2004;306(5703):1937–40.
Derks MF, Smit S, Salis L, Schijlen E, Bossers A, Mateman C, et al. The genome of winter moth (Operophtera brumata) provides a genomic perspective on sexual dimorphism and phenology. Genome Biol Evol. 2015;7(8):2321–32.
Nishikawa H, Iijima T, Kajitani R, Yamaguchi J, Ando T, Suzuki Y, et al. A genetic mechanism for female-limited Batesian mimicry in Papilio butterfly. Nat Genet. 2015;47(4):405.
Li X, Fan D, Zhang W, Liu G, Zhang L, Zhao L, et al. Outbred genome sequencing and CRISPR/Cas9 gene editing in butterflies. Nat Commun. 2015;6:8212.
Kanost MR, Arrese EL, Cao X, Chen Y-R, Chellapilla S, Goldsmith MR, et al. Multifaceted biological insights from a draft genome sequence of the tobacco hornworm moth, Manduca sexta. Insect Biochem Mol Biol. 2016;76:118–47.
Shen J, Cong Q, Kinch LN, Borek D, Otwinowski Z, Grishin NV. Complete genome of Pieris rapae, a resilient alien, a cabbage pest, and a source of anti-cancer proteins. F1000Res. 2016;5:2631.
Pearce SL, Clarke DF, East PD, Elfekih S, Gordon K, Jermiin LS, et al. Genomic innovations, transcriptional plasticity and gene loss underlying the evolution and divergence of two highly polyphagous and invasive Helicoverpa pest species. BMC Biol. 2017;15(1):63.
Cheng T, Wu J, Wu Y, Chilukuri RV, Huang L, Yamamoto K, et al. Genomic adaptation to polyphagy and insecticides in a major East Asian noctuid pest. Nat Ecol Evol. 2017;1(11):1747.
Wu N, Zhang S, Li X, Cao Y, Liu X, Wang Q, et al. Fall webworm genomes yield insights into rapid adaptation of invasive species. Nat Ecol Evol. 2019;3(1):105–15.
Zhang L, Liu B, Zheng W, Liu C, Zhang D, Zhao S, et al. High-depth resequencing reveals hybrid population and insecticide resistance characteristics of fall armyworm (Spodoptera frugiperda) invading China. bioRxiv. 2019:813154. https://doi.org/10.1101/813154.
Lu S, Yang J, Dai X, **e F, He J, Dong Z, et al. Chromosomal-level reference genome of Chinese peacock butterfly (Papilio bianor) based on third-generation DNA sequencing and Hi-C analysis. GigaScience. 2019;8(11):giz128.
Misof B, Liu S, Meusemann K, Peters RS, Donath A, Mayer C, et al. Phylogenomics resolves the timing and pattern of insect evolution. Science. 2014;346(6210):763–7.
Vogt RG, Große-Wilde E, Zhou J-J. The Lepidoptera odorant binding protein gene family: gene gain and loss within the GOBP/PBP complex of moths and butterflies. Insect Biochem Mol Biol. 2015;62:142–53.
Kristensen NP, Scoble MJ, Karsholt O. Lepidoptera phylogeny and systematics: the state of inventorying moth and butterfly diversity. Mol Phylogenet Evol. 2009;43(57):237–44.
Mutanen M, Wahlberg N, Kaila L. Comprehensive gene and taxon coverage elucidates radiation patterns in moths and butterflies. Proc Biol Sci. 2010;277(1695):2839–48.
Kawahara AY, Plotkin D, Espeland M, Meusemann K, Toussaint EF, Donath A, et al. Phylogenomics reveals the evolutionary timing and pattern of butterflies and moths. Proc Natl Acad Sci. 2019;116(45):22657–63.
Haines T, Horley D. Walking with beasts: a prehistoric safari: DK Pub; 2001.
Zhang L-W, Kang K, Jiang S-C, Zhang Y-N, Wang T-T, Zhang J, et al. Analysis of the antennal transcriptome and insights into olfactory genes in Hyphantria cunea (Drury). PLoS One. 2016;11(10):e0164729.
Faye I, Pye A, Rasmuson T, Boman HG, Boman I. Insect immunity. 11. Simultaneous induction of antibacterial activity and selection synthesis of some hemolymph proteins in diapausing pupae of Hyalophora cecropia and Samia cynthia. Infect Immun. 1975;12(6):1426–38.
Steiner H, Hultmark D, Engström Å, Bennich H, Boman H. Sequence and specificity of two antibacterial proteins involved in insect immunity. Nature. 1981;292(5820):246.
Gorman MJ, Paskewitz SM. Serine proteases as mediators of mosquito immune responses. Insect Biochem Mol Biol. 2001;31(3):257–62.
WIESNER A, LOSEN S, KOPÁČEK P, WEISE C, GÖTZ P. Isolated apolipophorin III from galleria mellonella stimulates the immune reactions of this insect. J Insect Physiol. 1997;43(4):383–91.
Kamita S, Maeda S. Inhibition of Bombyx mori nuclear polyhedrosis virus (NPV) replication by the putative DNA helicase gene of Autographa californica NPV. J Virol. 1993;67(10):6239–45.
Wu Q, Brown MR. Signaling and function of insulin-like peptides in insects. Annu Rev Entomol. 2006;51:1–24.
Riakhel A, Dhadialla T. Accumulation of yolk proteins in insects oocytes. Annu Rev Entomol. 1992;37:217–51.
Izumi S, Yano K, Yamamoto Y, Takahashi SY. Yolk proteins from insect eggs: structure, biosynthesis and programmed degradation during embryogenesis. J Insect Physiol. 1994;40(9):735–46.
Boman HG, Faye I, Gudmundsson GH, Lee JY, Lidholm DA. Cell-free immunity in Cecropia. A model system for antibacterial proteins. FEBS J. 2010;201(1):23–31.
Dolezelova E, Zurovec M, Dolezal T, Simek P, Bryant PJ. The emerging role of adenosine deaminases in insects. Insect Biochem Mol Biol. 2005;35(5):381–9.
Edgar BA. How flies get their size: genetics meets physiology. Nat Rev Genet. 2006;7(12):907.
Sap**ton TW, Raikhel AS. Molecular characteristics of insect vitellogenins and vitellogenin receptors. Insect Biochem Mol Biol. 1998;28(5–6):277–300.
Raubenheimer D, Simpson SJ. Integrative models of nutrient balancing: application to insects and vertebrates. Nutr Res Rev. 1997;10(1):151–79.
Raubenheimer D, Simpson SJ. Nutrient balancing in grasshoppers: behavioural and physiological correlates of dietary breadth. J Exp Biol. 2003;206(10):1669–81.
Feyereisen R, Koener JF, Cariño FA, Daggett AS. Biochemistry and Molecular Biology of Insect Cytochrome P450. US: Springer; 1990. p. 263–72.
Zhang L, Lu Y, **ang M, Shang Q, Gao X. The retardant effect of 2-Tridecanone, mediated by cytochrome P450, on the development of cotton bollworm, Helicoverpa armigera. BMC Genomics. 2016;17(1):954.
Niwa R, Matsuda T, Yoshiyama T, Namiki T, Mita K, Fujimoto Y, et al. CYP306A1, a cytochrome P450 enzyme, is essential for ecdysteroid biosynthesis in the prothoracic glands of Bombyx and Drosophila. J Biol Chem. 2004;279(34):35942–9.
Zhou H, Chen K, Yao Q, Gao L, Wang Y. Molecular cloning of Bombyx mori cytochrome P450 gene and its involvement in fluoride resistance. J Hazard Mater. 2008;160(2–3):330–6.
Nirmala X, ., Kodrík D, ., Zurovec M, ., Sehnal F, . Insect silk contains both a Kunitz-type and a unique Kazal-type proteinase inhibitor. FEBS J 2010;268(7):2064–2073.
Altman GH, Diaz F, Jakuba C, Calabro T, Horan RL, Chen J, et al. Silk-based biomaterials. Biomaterials. 2003;24(3):401–16.
Zhao P, Dong Z, Duan J, Wang G, Wang L, Li Y, et al. Genome-wide identification and immune response analysis of serine protease inhibitor genes in the silkworm, Bombyx mori. Plos One. 2012;7(2):e31168.
Yi Q, Zhao P, Wang X, Zou Y, Zhong X, Wang C, et al. Shotgun proteomic analysis of the Bombyx mori anterior silk gland: an insight into the biosynthetic fiber spinning process. Proteomics. 2013;13(17):2657–63.
Inoue S, Tanaka K, Arisaka F, Kimura S, Ohtomo K, Mizuno S. Silk fibroin of Bombyx mori is secreted, assembling a high molecular mass elementary unit consisting of H-chain, L-chain, and P25, with a 6:6:1 molar ratio. J Biol Chem. 2000;275(51):40517–28.
Sutherland TD, Young JH, Weisman S, Hayashi CY, Merritt DJ. Insect silk: one name, many materials. Annu Rev Entomol. 2010;55(1):171.
Song F, Zhang P, Yi F, Hong X, Lu C, Yutaka B, et al. Study on fibroin heavy chain of the silkworm Bombyx mori by fluorescence in situ hybridization (FISH). Sci China. 2002;45(6):663–8.
Wang X, Fang X, Yang P, Jiang X, Jiang F, Zhao D, et al. The locust genome provides insight into swarm formation and long-distance flight. Nat Commun. 2014;5(5):2957.
Miller KG, Browning JV, Aubry MP, Wade BS, Katz ME, Kulpecz AA, et al. Eocene-Oligocene global climate and sea-level changes: St. Stephens quarry, Alabama. Geol Soc Am Bull. 2008;120(1):34–53.
Wolfe JA. A Paleobotanical interpretation of tertiary climates in the northern hemisphere: data from fossil plants make it possible to reconstruct tertiary climatic changes, which may be correlated with changes in the inclination of the earth's rotational axis. Am Sci. 1978;66(6):694–703.
Berggren WA, Prothero DR. Eocene-Oligocene climatic and biotic evolution. Princeton: Princeton University Press; 1992.
Mei T, Fu W-B, Li B, He Z-B, Chen B. Comparative genomics of chemosensory protein genes (CSPs) in twenty-two mosquito species (Diptera: Culicidae): identification, characterization, and evolution. PLoS One. 2018;13(1):e0190412.
Xu W, Alexie P, Zhang HJ, Alisha A. Expansion of a bitter taste receptor family in a polyphagous insect herbivore. Sci Rep. 2016;6:23666.
Li X, Schuler MA, Berenbaum MR. Molecular mechanisms of metabolic resistance to synthetic and natural xenobiotics. Annu Rev Entomol. 2007;52(1):231.
Tsubota T, Shiotsuki T. Genomic and phylogenetic analysis of insect carboxyl/cholinesterase genes. J Pestic Sci. 2010;35(2):310–4.
Ahn S, Vogel H, Heckel D. Comparative analysis of the UDP-glycosyltransferase multigene family in insects. Insect Biochem Mol Biol. 2012;42(2):133–47.
Zhou D, Liu X, Sun Y, Ma L, Shen B, Zhu C. Genomic analysis of detoxification supergene families in the mosquito Anopheles sinensis. PLoS One. 2015;10(11):e0143387.
Claudianos C, Ranson H, Johnson R, Biswas S, Schuler M, Berenbaum M, et al. A deficit of detoxification enzymes: pesticide sensitivity and environmental response in the honeybee. Insect Mol Biol. 2006;15(5):615–36.
Duman JG, Xu L, Neven LG, Tursman D, Wu DW. Hemolymph proteins involved in insect subzero-temperature tolerance: ice nucleators and antifreeze proteins. Insects at low temperature: Springer; 1991. p. 94–127.
Andreeva-Kovalevskaya ZI, Solonin A, Sineva E, Ternovsky V. Pore-forming proteins and adaptation of living organisms to environmental conditions. Biochem Mosc. 2008;73(13):1473–92.
Srinivasan A, Giri AP, Gupta VS. Structural and functional diversities in lepidopteran serine proteases. Cell Mol Biol Lett. 2006;11(1):132.
Wang J, Li DZ, Min SF, Mi F, Zhou SS, Wang MQ. Analysis of chemosensory gene families in the beetle Monochamus alternatus and its parasitoid Dastarcus helophoroides. Comp Biochem Physiol Part D Genomics Proteomics. 2014;11(9):1–8.
do ARB N, Fresia P, Cônsoli FL, Omoto C. Comparative transcriptome analysis of lufenuron-resistant and susceptible strains of Spodoptera frugiperda (Lepidoptera: Noctuidae). BMC Genomics. 2015;16(1):985.
Yu QY, Fang SM, Zhang Z, Jiggins CD. The transcriptome response of Heliconius melpomene larvae to a novel host plant. Mol Ecol. 2016;25(19):4850–65.
Dermauw W, Wybouw N, Rombauts S, Menten B, Vontas J, Grbić M, et al. A link between host plant adaptation and pesticide resistance in the polyphagous spider mite Tetranychus urticae. Proc Natl Acad Sci. 2013;110(2):E113–E22.
Strode C, Steen K, Ortelli F, Ranson H. Differential expression of the detoxification genes in the different life stages of the malaria vector Anopheles gambiae. Insect Mol Biol. 2006;15(4):523–30.
Rey D, Cuany A, Pautou M-P, Meyran J-C. Differential sensitivity of mosquito taxa to vegetable tannins. J Chem Ecol. 1999;25(3):537–48.
Hennigesjanssen K, Reineke A, Heckel DG, Groot AT. Complex inheritance of larval adaptation in Plutella xylostella to a novel host plant. Heredity. 2011;107(5):421.
JJMe X. Invited review: microbial ecology in the age of genomics and metagenomics: concepts, tools, and recent advances. Mol Ecol. 2006;15(7):1713–31.
Shi W, Syrenne R, Sun JZ, JSJIS Y. Molecular approaches to study the insect gut symbiotic microbiota at the ‘omics’ age. Insect Science. 2010;17(3):199–219.
Colman DR, Toolson EC, CJME T-V. Do diet and taxonomy influence insect gut bacterial communities? Mol Ecol. 2012;21(20):5124–37.
**ang H, Wei G-F, Jia S, Huang J, Miao X-X, Zhou Z, et al. Microbial communities in the larval midgut of laboratory and field populations of cotton bollworm (Helicoverpa armigera). Can J Microbiol. 2006;52(11):1085–92.
**ang H, Li M, Zhao Y, Zhao L, Zhang Y, Huang Y. Bacterial community in midguts of the silkworm larvae estimated by PCR/DGGE and 16S rDNA gene library analysis; 2007.
Delalibera I Jr, Handelsman J, KFJEE R. Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environ Entomol. 2005;34(3):541–7.
Dixon R, DJNRM K. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2(8):621.
Behar A, Yuval B, EJME J. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol. 2005;14(9):2637–43.
Delalibera I Jr, Handelsman J, Raffa KF. Contrasts in cellulolytic activities of gut microorganisms between the wood borer, Saperda vestita (Coleoptera: Cerambycidae), and the bark beetles, Ips pini and Dendroctonus frontalis (Coleoptera: Curculionidae). Environ Entomol. 2005;34(3):541–7.
Dixon R, Kahn D. Genetic regulation of biological nitrogen fixation. Nat Rev Microbiol. 2004;2(8):621.
Behar A, Yuval B, Jurkevitch E. Enterobacteria-mediated nitrogen fixation in natural populations of the fruit fly Ceratitis capitata. Mol Ecol. 2005;14(9):2637–43.
Dillon RJ, Dillon VM. The gut bacteria of insects: nonpathogenic interactions. Annu Rev Entomol. 2004;49(1):71–92 PubMed PMID: 14651457. Epub 2003/12/04.
Chen B, Yu T, **e S, Du K, Liang X, Lan Y, et al. Comparative shotgun metagenomic data of the silkworm Bombyx mori gut microbiome. Sci Data. 2018;5:180285.
Suen G, Scott JJ, Aylward FO, Adams SM, Tringe SG, Pinto-Tomas AA, et al. An insect herbivore microbiome with high plant biomass-degrading capacity. PLoS Genet. 2010;6(9):e1001129 PubMed PMID: 20885794. Pubmed Central PMCID: PMC2944797. Epub 2010/10/05.
Pinto-Tomas AA, Anderson MA, Suen G, Stevenson DM, FST C, Cleland WW, et al. Symbiotic Nitrogen Fixation in the Fungus Gardens of Leaf-Cutter Ants. Science. 2009;326(5956):1120–3 PubMed PMID: WOS:000271951000047. English.
Yadav AN, Sharma D, Gulati S, Singh S, Dey R, Pal KK, et al. Haloarchaea endowed with phosphorus solubilization attribute implicated in phosphorus cycle. Sci Rep. 2015;5:12293.
Yadav AN, Verma P, Kaushik R, Dhaliwal H, AJEM S. Archaea endowed with plant growth promoting attributes. EC Microbiol. 2017;8(6):294–8.
Kikuchi Y, Hosokawa T, TJA F. Insect-microbe mutualism without vertical transmission: a stinkbug acquires a beneficial gut symbiont from the environment every generation. Appl Environ Microbiol. 2007;73(13):4308–16.
Kikuchi Y, Hayatsu M, Hosokawa T, Nagayama A, Tago K, Fukatsu T. Symbiont-mediated insecticide resistance. Proc Natl Acad Sci U S A. 2012;109(22):8618–22.
Keeling CI, JJNP B. Genes, enzymes and chemicals of terpenoid diversity in the constitutive and induced defence of conifers against insects and pathogens. New Phytol. 2006;170(4):657–75.
Minard G, Mavingui P, Moro CV. Diversity and function of bacterial microbiota in the mosquito holobiont. Parasit Vectors. 2013;6(1):146.
**a X, Gurr GM, Vasseur L, Zheng D, Zhong H, Qin B, et al. Metagenomic sequencing of diamondback moth gut microbiome unveils key holobiont adaptations for herbivory. Front Microbiol. 2017;8:663.
Sticklen MB. Plant genetic engineering for biofuel production: towards affordable cellulosic ethanol. Nat Rev Genet. 2008;9(6):433.
Liu N, Zhang L, Zhou H, Zhang M, Yan X, Wang Q, et al. Metagenomic insights into metabolic capacities of the gut microbiota in a fungus-cultivating termite (Odontotermes yunnanensis). PLoS One. 2013;8(7):e69184.
Patel DD, Patel AK, Parmar NR, Shah TM, Patel JB, Pandya PR, et al. Microbial and Carbohydrate Active Enzyme profile of buffalo rumen metagenome and their alteration in response to variation in the diet. Gene. 2014;545(1):88–94.
USA MDoC. Fall webworm Hyphantria cunea (Drury). External Factsheets. 2000.
Fitzgerald TD. Sociality in caterpillar; 1993.
Takuya T, Kimiko Y, Kazuei M, et al. Gene expression analysis in the larval silk gland of the eri silkworm Samia ricini. Insect Sci. 2016;23(6):791–804.
Rehnberg BG. Temperature profiles inside webs of the fall webworm, Hyphantria cunea (Lepidoptera: Arctiidae): Influence of weather, compass orientation, and time of day. J Therm Biol. 2006;31(3):274–9.
Mori H, Tsukada M. New silk protein: modification of silk protein by gene engineering for production of biomaterials. Rev Mol Biotechnol. 2000;74(2):95–103.
Kato N, Sato S, Yamanaka A, Yamada H, Fuwa N, Nomura M. Silk protein, sericin, inhibits lipid peroxidation and tyrosinase activity. J Agr Chem Soc Jpn. 1998;62(1):145–7.
Terada S, Nishimura T, Sasaki M, Yamada H, Miki M. Sericin, a protein derived from silkworms, accelerates the proliferation of several mammalian cell lines including a hybridoma. Cytotechnology. 2002;40(1–3):3–12.
Terada S, Sasaki M, Yanagihara K, Yamada H. Preparation of silk protein sericin as mitogenic factor for better mammalian cell culture. J Biosci Bioeng. 2005;100(6):667–71.
Morikawa M, Kimura T, Murakami M, Katayama K, Terada S, Yamaguchi A. Rat islet culture in serum-free medium containing silk protein sericin. J Hepatobiliary Pancreatic Surg. 2009;16(2):223–8.
Manosroi A, Boonpisuttinant K, Winitchai S, Manosroi W, Manosroi J. Free radical scavenging and tyrosinase inhibition activity of oils and sericin extracted from Thai native silkworms (Bombyx mori). Pharm Biol. 2010;48(8):855–60.
Yang M. Silk-based biomaterials. Microsc Res Tech. 2017;80(3):321–30.
Li S, Liu B, Cheng J, Hu J. Composite cement of magnesium-bearing phosphoaluminate–hydroxyapatite reinforced by treated raw silk fiber. Cement Concrete Composites. 2008;30(4):347–52.
Oyama F, Mizuno S, Shimura K. Studies on immunological properties of fibroin heavy and light chains. J Biochem. 1984;96(6):1689–94.
Matsuno K, ., Hui CC, Takiya S, ., Suzuki T, ., Ueno K, ., Suzuki Y, . Transcription signals and protein binding sites for sericin gene transcription in vitro. J Biol Chem 1989;264(31):18707–18713.
Matsuno K, Takiya S, Hui CC, Suzuki T, Fukuta M, Ueno K, et al. Transcriptional stimulation via SC site of Bombyx sericin-1 gene through an interaction with a DNA binding protein SGF-3. Nucleic Acids Res. 1990;18(7):1853–8.
Ohno K, Sawada JI, Takiya S, Mai K, Matsumoto A, Tsubota T, et al. Silk Gland Factor-2 (SGF-2) Involved in Fibroin Gene Transcription Consists of LIM-homeodomain, LIM-interacting, and Single-Stranded DNA-Binding Proteins. J Biol Chem. 2013;288(44):31581.
Tsuda M, ., Suzuki Y, . Faithful transcription initiation of fibroin gene in a homologous cell-free system reveals an enhancing effect of 5′ flanking sequence far upstream. Cell 1981;27(1):175–182.
Takiya S, ., Hui CC, Suzuki Y, . A contribution of the core-promoter and its surrounding regions to the preferential transcription of the fibroin gene in posterior silk gland extracts. Embo J. 1990;9(2):489–496.
Guo PC, Dong Z, Zhao P, Zhang Y, He H, Tan X, et al. Structural insights into the unique inhibitory mechanism of the silkworm protease inhibitor serpin18. Sci Rep. 2015;55:11863.
Zhang Y, Zhao P, Dong Z, Wang D, Guo P, Guo X, et al. Comparative proteome analysis of multi-layer cocoon of the silkworm, Bombyx mori. Plos One. 2015;10(4):e0123403.
Biosciences P. Procedure & Checklist—20 kb Template Preparation Using BluePippinTM Size Selection System. SampleNet; 2014.
Hale CM, Chen W-C, Khatau SB, Daniels BR, Lee JS, Wirtz D. SMRT analysis of MTOC and nuclear positioning reveals the role of EB1 and LIC1 in single-cell polarization. J Cell Sci. 2011;124(Pt 24):4267–85.
Koren S, Walenz BP, Berlin K, Miller JR, Bergman NH, Phillippy AM. Canu: scalable and accurate long-read assembly via adaptive k-mer weighting and repeat separation. Genome Res. 2017;27(5):722–36.
Shigang W. A fuzzy Bruijn graph approach to long noisy reads assembly 2017 [cited 2018 12th,oct]. Available from: https://github.com/ruanjue/wtdbg.
De Landtsheer S, Trairatphisan P, Lucarelli P, TJB S. FALCON: a toolbox for the fast contextualization of logical networks. Bioinformatics. 2017;33(21):3431–6.
Walker BJ, Abeel T, Shea T, Priest M, Abouelliel A, Sakthikumar S, et al. Pilon: an integrated tool for comprehensive microbial variant detection and genome assembly improvement. PLoS One. 2014;9(11):e112963 PubMed PMID: 25409509. Pubmed Central PMCID: PMC4237348. Epub 2014/11/20.
Parra G, Bradnam K, IJB K. CEGMA: a pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics. 2007;23(9):1061–7.
Simão FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM, et al. Bioinformatics. 2015;31(19):3210–2.
Xu Z, HJNar W. LTR_FINDER: an efficient tool for the prediction of full-length LTR retrotransposons. Nucleic Acids Res. 2007;35(suppl_2):W265–W8.
Han Y, Wessler SR. MITE-Hunter: a program for discovering miniature inverted-repeat transposable elements from genomic sequences. Nucleic Acids Res. 2010;38(22):e199.
Price AL, Jones NC, Pevzner PAJB. De novo identification of repeat families in large genomes. Bioinformatics. 2005;21(suppl_1):i351–i8.
Edgar RC, Myers EWJB. PILER: identification and classification of genomic repeats. Bioinformatics. 2005;21(suppl_1):i152–i8.
Wicker T, Sabot F, Hua-Van A, Bennetzen JL, Capy P, Chalhoub B, et al. A unified classification system for eukaryotic transposable elements. Nat Rev Genet. 2007;8(12):973.
Chen N. Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics. 2004;5(1):4.10 1–4.. 4.
Lowe TM. tRNAscan-SE: a program for improved detection of transfer RNA genes in genomic sequence. Eddy SR. 1997;25(5):955.
Nawrocki EP, Eddy SRJB. Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics. 2013;29(22):2933–5.
She R, Chu JS-C, Wang K, Pei J, Chen N. GenBlastA: enabling BLAST to identify homologous gene sequences. Genome Res. 2009;19(1):143–9.
Birney E, Clamp M, Durbin R. GeneWise and genomewise. Genome Res. 2004;14(5):988–95.
Burge C, Karlin S. Prediction of complete gene structures in human genomic DNA1. J Mol Biol. 1997;268(1):78–94.
Stanke M, Waack SJB. Gene prediction with a hidden Markov model and a new intron submodel. Bioinformatics. 2003;19(suppl_2):ii215–i25.
Majoros WH, Pertea M, Salzberg SL. TigrScan and GlimmerHMM: two open source ab initio eukaryotic gene-finders. Bioinformatics. 2004;20(16):2878–9.
Blanco E, Parra G, Guigó R. Using geneid to identify genes. Curr Protoc Bioinformatics. 2007;18(1):4.3 1–4.3. 28.
Korf I. Gene finding in novel genomes. BMC Bioinformatics. 2004;5(1):59.
Keilwagen J, Wenk M, Erickson JL, Schattat MH, Grau J, Hartung F. Using intron position conservation for homology-based gene prediction. Nucleic Acids Res. 2016;44(9):e89.
Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript-level expression analysis of RNA-seq experiments with HISAT, StringTie and Ballgown. Nat Protoc. 2016;11(9):1650.
Haas B, Papanicolaou AJGS. TransDecoder (find coding regions within transcripts); 2016.
Tang S, Lomsadze A, Borodovsky M. Identification of protein coding regions in RNA transcripts. Nucleic Acids Res. 2015;43(12):e78.
Campbell MA, Haas BJ, Hamilton JP, Mount SM, Buell CR. Comprehensive analysis of alternative splicing in rice and comparative analyses with Arabidopsis. BMC Genomics. 2006;7(1):327.
Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, et al. Automated eukaryotic gene structure annotation using EVidenceModeler and the Program to Assemble Spliced Alignments. Genome Biol. 2008;9(1):1.
Chen F, Mackey AJ, Stoeckert CJ Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34(suppl_1):D363–D8.
Löytynoja A, Goldman N. An algorithm for progressive multiple alignment of sequences with insertions. Proc Natl Acad Sci U S A. 2005;102(30):10557–62.
Castresana J. Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol Biol Evol. 2000;17(4):540–52.
Shao Y, Li J-X, Ge R-L, Zhong L, Irwin DM, Murphy RW, et al. Genetic adaptations of the plateau zokor in high-elevation burrows. Sci Rep. 2015;5:17262.
Stamatakis AJB. RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics. 2014;30(9):1312–3.
Sanderson MJJB. r8s: inferring absolute rates of molecular evolution and divergence times in the absence of a molecular clock. Bioinformatics. 2003;19(2):301–2.
Durden CJ, Rose H. Butterflies from the middle Eocene: the earliest occurrence of fossil Papilionoidea (Lepidoptera). Texas Memorial Museum, The University of Texas at Austin; 1978.
Lukashevich ED, Przhiboro AA, Marchal-Papier F, Grauvogel-Stamm L. The oldest occurrence of immature Diptera (Insecta). Middle Triassic, France: Annales de la Société entomologique de France, Taylor & Francis Group. 2010;46(1-2):4-22.
Kirejtshuk AG, Poschmann M, Prokop J, Garrouste R, Nel A. Evolution of the elytral venation and structural adaptations in the oldest Palaeozoic beetles (Insecta: Coleoptera: Tshekardocoleidae). J Syst Palaeontology. 2014;12(5):575–600.
De Bie T, Cristianini N, Demuth JP, Hahn MW. CAFE: a computational tool for the study of gene family evolution. Bioinformatics. 2006;22(10):1269–71.
Yang Z. PAML 4: phylogenetic analysis by maximum likelihood. Mol Biol Evol. 2007;24(8):1586–91.
Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods. 2015;12(4):357.
Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, Van Baren MJ, et al. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nature Biotechnol. 2010;28(5):511.
Robinson MD, DJ MC, Smyth GK. edgeR: a Bioconductor package for differential expression analysis of digital gene expression data. Bioinformatics. 2010;26(1):139–40.
Leng N, Dawson JA, Thomson JA, Ruotti V, Rissman AI, Smits BM, et al. EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics. 2013;29(8):1035–43.
Haynes W. Benjamini–hochberg method. Encyclopedia of systems biology; 2013. p. 78.
Wood DE, Salzberg SL. Kraken: ultrafast metagenomic sequence classification using exact alignments. Genome Biol. 2014;15(3):R46.
Ondov BD, Bergman NH, Phillippy AM. Genomes, Metagenomes: Basics M, Databases, Tools. Krona: Interactive Metagenomic Visualization in a Web Browser; 2015. p. 339–46.
Peng Y, Leung HC, Yiu S-M, Chin FY. IDBA-UD: a de novo assembler for single-cell and metagenomic sequencing data with highly uneven depth. Bioinformatics. 2012;28(11):1420–8.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–5.
Zhu W, Lomsadze A, Borodovsky M. Ab initio gene identification in metagenomic sequences. Nucleic Acids Res. 2010;38(12):e132.
Fu L, Niu B, Zhu Z, Wu S, Li W. CD-HIT: accelerated for clustering the next-generation sequencing data. Bioinformatics. 2012;28(23):3150–2.
Finn RD, Clements J, Arndt W, Miller BL, Wheeler TJ, Schreiber F, et al. HMMER web server: 2015 update. Nucleic Acids Res. 2015;43(W1):W30–W8.
Acknowledgements
We appreciate the critical editing and proofreading of JJ Scientific Consultant Ltd., UK, and American Journal Experts.
Funding
Funding for this study came from the National Natural Science Foundation of China (No. 31501890, YLW), the Natural Science Foundation of Jilin Province (No. 20160204023NY and No. 20170204003NY, BZR; No. 20180101005JC and No. 20190301047NY, YLW), the Foundation of **njiang Uygur Autonomous Region (No. 2017E0272), the Open Project Program of the Jilin Provincial Key Laboratory of Animal Resource Conservation and Utilization (No. 130028684 and No. 1300289103), the Fundamental Research Funds for the Central Universities (No. 11SSXT153, No. 2412015KJ015 and No. 2412019FZ022), the Fund for Fostering Talents in Basic Science of the National Natural Science (No. J1210070), and the Undergraduate teaching quality and teaching reform project of Northeast Normal University (No. 131004003). The funding bodies played no role in writing the manuscript.
Author information
Authors and Affiliations
Contributions
Qi Chen and Hanbo Zhao contributed equally to this work. YLW and BZR designed the project; Biomarker Technologies Corporation performed the sequencing, assembly and functional annotation work; QC, HBZ and JXL performed the data analyses; JXL, JTW, MW, HFZ and YXZ performed the experiments; QC, HBZ, YLW and JJZ wrote the manuscript with input from all the other authors, and all authors read and approved the final manuscript version.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
Not applicable.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Additional file 1: Figure S1
. K-mer distribution of preprocessed data with k = 19. The distribution of depth analysis based on whole genome data in the fall webworm. Using the formula: genome size = k-mer count/peak of the kmer distribution, thereinto, k = 19.
Additional file 2: Figure S2.
Numbers of genes annotated with three gene prediction strategies. The final number of genes supported by homologous prediction and transcriptome prediction was 14,688, accounting for a significant proportion (95.88%) of 15,319 (the total number of protein-coding genes), showing the high quality of the prediction.
Additional file 3: Figure S3.
GO annotation of the H. cunea genome. The capital letters on the x-axis indicate the GO categories as listed below, the left y-axis indicates the percentage of genes in each category, and the right y-axis indicates the number of genes in each category.
Additional file 4: Figure S4.
KOG annotation of the H. cunea genome. The capital letters on the x-axis indicate the KOG classification as listed on the right, and the y-axis indicates the number of genes in each classification.
Additional file 5: Figure S5.
The distribution of Nr homologous genes within the H. cunea genome in insect species. The percentage of Nr homologous genes over the H. cunea genome were obtained by EVidenceModeler (EVM) with more ten insect species, including Bombyx mori, Danaus plexippus, Helicoverpa armigera, Papilio xuthus, Manduca sexta, Spodoptera frugiperda, Papilio polytes, Tribolium castaneum, Spodoptera litura and Spodoptera exigua.
Additional file 6 Figure S6.
Box plot of FPKM values from different developmental stages and tissues. Box plot of log10 FPKM values aggregated across the 8232 DEGs of the stage RNA sequencing groups and 7733 DEGs of the tissue RNA sequencing groups.
Additional file 7: Figure S7.
Numbers of alternative splicing events in different tissues and stages of H. cunea. The horizontal axis represents the number of alternative splicing events under the corresponding event, and the vertical axis represents the abbreviation of the classification of alternative splicing events. (1) AE: Alternative exon ends; (2) IR: Intron retention (IR_ON, IR_OFF pair); (3) MIR: Multi-IR (MIR_ON, MIR_OFF pair); (4) MSKIP: Multiexon SKIP (MSKIP ON, MSKIP OFF pair); (5) SKIP: Skipped exon (SKIP ON, SKIP OFF pair); (6) TSS: Alternative 5′ first exon (transcription start site); (7) TTS: Alternative 3′ last exon (transcription terminal site); (8) XAE: Approximate AE; (9) XIR: Approximate IR (XIR ON, XIR OFF pair); (10) XMIR: Approximate MIR (XMIR ON, XMIR OFF pair); (11) XMSKIP: Approximate MSKIP (XMSKIP ON, XMSKIP OFF pair); (12) XSKIP: Approximate SKIP (XSKIP ON, XSKIP OFF pair).
Additional file 8: Figure S8.
COG functional enrichment analysis annotation of the metagenomic data. L, R, C, G and O are the top five gene function categories. The remaining categories were defined as “Others” which including N: cell motility; D: Cell cycle control, cell division, chromosome partitioning; A: RNA processing and moditication; M: Cell wall/membrance/envelope biogenesis; U: Intracellular trafficking,secretion, and vesicular transport.
Additional file 9: Figure S9.
Proportion of Carbohydrate-Active EnZymes in the metagenome data. BLASTp was used to compare the sequences of the nonredundant gene sets with the CAZy database to obtain the gene annotation information.
Additional file 10: Table S1.
Results of the preliminary survey. Table S2. Prediction of noncoding genes. Table S3. Statistical information of two-algebra data by Burrow-Wheeler Aligner (BWA). Table S4. Integrity evaluation of Illumina data. Table S5. Integrity evaluation of PacBio data by CEGMA. Table S6. Integrity evaluation of PacBio data by BUSCO. Table S7. DEG statistical results from different stages and tissues. Table S8. Sequencing statistics of metagenomic data. Table S13. The detail of the genome versions used in this study. Table S14. Primers used for RNA interference and RT-qPCR.
Additional file 11: Table S9.
Annotation results of the metagenome data. Table S10. KEGG enrichment analysis of metagenomic genes. Table S11. Metagenomic enzymes associated with cellulose and hemicellulose hydrolysis. Table S12. Metagenomic gene sets encoding glutathione S-transferase, esterases, and P450s.
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated in a credit line to the data.
About this article
Cite this article
Chen, Q., Zhao, H., Wen, M. et al. Genome of the webworm Hyphantria cunea unveils genetic adaptations supporting its rapid invasion and spread. BMC Genomics 21, 242 (2020). https://doi.org/10.1186/s12864-020-6629-6
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-020-6629-6