Abstract
Background
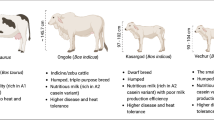
Mithun (Bos frontalis), also called gayal, is an endangered bovine species, under the tribe bovini with 2n = 58 XX chromosome complements and reared under the tropical rain forests region of India, China, Myanmar, Bhutan and Bangladesh. However, the origin of this species is still disputed and information on its genomic architecture is scanty so far. We trust that availability of its whole genome sequence data and assembly will greatly solve this problem and help to generate many information including phylogenetic status of mithun. Recently, the first genome assembly of gayal, mithun of Chinese origin, was published. However, an improved reference genome assembly would still benefit in understanding genetic variation in mithun populations reared under diverse geographical locations and for building a superior consensus assembly. We, therefore, performed deep sequencing of the genome of an adult female mithun from India, assembled and annotated its genome and performed extensive bioinformatic analyses to produce a superior de novo genome assembly of mithun.
Results
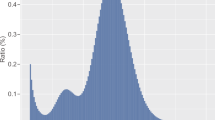
We generated ≈300 Gigabyte (Gb) raw reads from whole-genome deep sequencing platforms and assembled the sequence data using a hybrid assembly strategy to create a high quality de novo assembly of mithun with 96% recovered as per BUSCO analysis. The final genome assembly has a total length of 3.0 Gb, contains 5,015 scaffolds with an N50 value of 1 Mb. Repeat sequences constitute around 43.66% of the assembly. The genomic alignments between mithun to cattle showed that their genomes, as expected, are highly conserved. Gene annotation identified 28,044 protein-coding genes presented in mithun genome. The gene orthologous groups of mithun showed a high degree of similarity in comparison with other species, while fewer mithun specific coding sequences were found compared to those in cattle.
Conclusion
Here we presented the first de novo draft genome assembly of Indian mithun having better coverage, less fragmented, better annotated, and constitutes a reasonably complete assembly compared to the previously published gayal genome. This comprehensive assembly unravelled the genomic architecture of mithun to a great extent and will provide a reference genome assembly to research community to elucidate the evolutionary history of mithun across its distinct geographical locations.
Similar content being viewed by others
Background
Mithun (Bos frontalis) is a rare bovine species living under free-range conditions inside tropical rainforest ecosystems of India, Bangladesh, Bhutan, China, and Myanmar [1]. It is a unique animal having a massive body, with characteristic ‘white stockings’ on their stout legs. This animal efficiently converts grass, forage, tree leaves as well as various agricultural by-products into highly nutritious meat. Moreover, mithun holds a unique place in the evolution of bovines. Mithun, having a specific chromosomal pattern, 2n = 58 is distinguishable from that of cattle (2n = 60) and yak (2n = 60) [2]. However, the origin of mithun is an on-going debate with no well-supported conclusion [3,4,59] flagstat were used to investigate the correctness of assembly. BUSCO v3 [20, 60] with the mammalian database was used to assess the completeness of genes presented by assembly. Nineteen-mers was counted from PE data with Jellyfish [61]. Genome size was estimated by dividing the total number of k-mers by the peak value of the k-mer frequency distribution [77] pathway information of [77] pathway information of the mithun gene set.
Genome alignment
Both mithun and gayal genomes were soft-masked and aligned to the soft-masked cattle genome (ARS-UCD1.2) [21] by Large Scale Genome Alignment Tools (LASTZ) [78]. The pairwise genome alignment was chained according to their location in both genomes by axtChain program [79]. The netting process chooses for the reference species the best sub-chain in each region. The statistics of different size of synteny block was done by a custom script. We only used block size larger than 100 kb to investigate how many cattle chromosomes the mithun scaffold can span.
Availability of data and materials
All data generated and analyzed during this current study are available in the Data Cell of ICAR-NRC on Mithun, Nagaland, India with permission from the Competent Authority. Illumina whole genome sequence and RNA-seqdata of mithun were submitted in NCBI Database having BioProject ID PRJNA241403 and BioProject ID PRJNA307305, respectively.
Abbreviations
- BTA 7:
-
Bos taurus genome assembly v.7
- BUSCO:
-
Benchmarking Universal Single-Copy Orthologs
- CNVs:
-
Copy number variations
- DBG:
-
De Bruijn Graphs
- dNTPs:
-
Deoxyribonucleotide triphosphates
- eggNOG:
-
Evolutionary genealogy of genes: Non-supervised Orthologous Groups
- EVM:
-
EVidence Modeler
- Gb:
-
Gigabyte
- KAAS:
-
KEGG Automatic Annotation Server
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- LASTZ:
-
Large-Scale Genome Alignment Tool
- LINEs:
-
Long Interspersed Nuclear Elements
- LoRDEC:
-
Long Read DBG Error Correction
- LTR:
-
Long terminal repeat
- MaSuRCA:
-
Maryland Super-Read Celera Assembler
- MB:
-
Megabyte
- Mt.:
-
Mitochondria
- NGS:
-
Next-generation sequencing
- PacBio:
-
Pacific BioSciences
- PASApipeline:
-
Program to Assemble Spliced Alignments Pipeline
- PBJelly 2:
-
Software for Long-Read Sequencing Data from PacBio
- Pfam:
-
Protein families database
- PRINTS:
-
Protein fingerprints database
- QUAST:
-
Quality Assessment Tools
- RIN:
-
RNA Integrity Number
- SINEs:
-
Short Interspersed Nuclear Elements
- SMRT:
-
Single-molecule real-time (SMRT)
- SNPs:
-
Single-Nucleotide Polymorphisms
- SSPACE:
-
Scaffolding Pre-Assemblies AfterContig Extension
- SVs:
-
Structural variations
References
Mondal M, Dhali A, Rajkhowa C, Prakash BS. Secretion patterns of growth hormone in growing captive mithuns (Bos frontalis). Zool Sci. 2004;21(11):1125–9.
Gupta N, Verma ND, Gupta SC, Kumar P, Sahai R. Chromosomes of Mithun (Gaveaus-frontalis). Indian J An Sci. 1995;65(6):688–90.
FJaS S, Elizabeth S. a ceremonial ox of India: the mithun in nature, culture, and history, with notes on the domestication of common cattle. Madison, WI: The University of Wisconsin Press; 1968.
Tanaka K, Takizawa T, Murakoshi H, Dorji T, Nyunt MM, Maeda Y, Yamamoto Y, Namikawa T. Molecular phylogeny and diversity of Myanmar and Bhutan mithun based on mtDNA sequences. Anim Sci J. 2011;82(1):52–6.
Lan H, **ong X, Lin S, Liu A, Shi L. Mitochondrial DNA polymorphism of cattle (Bos taurus) and mithun (Bos frontalis) in Yunnan Province. Yi chuan xue bao=. Acta Genet Sin. 1992;20(5):419–25.
Winter H, Mayr B, Schleger W, Dworak E, Krutzler J, Kalat M. Genetic characterisation of the mithun (Bos frontalis) and studies of spermatogenesis, blood groups and haemoglobins of its hybrids with Bos indicus. Res Vet Sci. 1986;40(1):8–17.
Mei C, Wang H, Zhu W, Wang H, Cheng G, Qu K, Guang X, Li A, Zhao C, Yang W, et al. Whole-genome sequencing of the endangered bovine species Gayal (Bos frontalis) provides new insights into its genetic features. Sci Rep. 2016;6:19787.
Wang MS, Zeng Y, Wang X, Nie WH, Wang JH, Su WT, Otecko NO, **ong ZJ, Wang S, Qu KX, et al. Draft genome of the gayal. Bos frontalis Gigascience. 2017;6(11):1–7.
Bovine Genome S, Analysis C, Elsik CG, Tellam RL, Worley KC, Gibbs RA, Muzny DM, Weinstock GM, Adelson DL, Eichler EE, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324(5926):522–8.
Weldenegodguad M, Popov R, Pokharel K, Ammosov I, Ming Y, Ivanova Z and Kantanen J. 2019. Whole-Genome Sequencing of Three Native Cattle Breeds Originating From the Northernmost Cattle Farming Regions. Front. Genet. Published online 2019. https://doi.org/10.3389/fgene.2018.00728.
Tantia MS, Vijh RK, Bhasin V, Sikka P, Vij PK, Kataria RS, Mishra BP, Yadav SP, Pandey AK, Sethi RK, et al. Whole-genome sequence assembly of the water buffalo (Bubalus bubalis). Indian J Animal Sci. 2011;81(5):465–73.
Qiu Q, Zhang G, Ma T, Qian W, Wang J, Ye Z, Cao C, Hu Q, Kim J, Larkin DM, et al. The yak genome and adaptation to life at high altitude. Nat Genet. 2012;44(8):946–9.
McCoy RC, Taylor RW, Blauwkamp TA, Kelley JL, Kertesz M, Pushkarev D, Petrov DA, Fiston-Lavier AS. Illumina TruSeq synthetic long-reads empower de novo assembly and resolve complex, highly-repetitive transposable elements. PLoS One. 2014;9(9):e106689.
Caporaso JG, Lauber CL, Walters WA, Berg-Lyons D, Huntley J, Fierer N, Owens SM, Betley J, Fraser L, Bauer M, et al. Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms. ISME J. 2012;6(8):1621–4.
Rhoads A, Au KF. PacBio sequencing and its applications. Genomics Proteomics Bioinformatics. 2015;13(5):278–89.
Zimin AV, Marcais G, Puiu D, Roberts M, Salzberg SL, Yorke JA. The MaSuRCA genome assembler. Bioinformatics. 2013;29(21):2669–77.
Boetzer M, Henkel CV, Jansen HJ, Butler D, Pirovano W. Scaffolding pre-assembled contigs using SSPACE. Bioinformatics. 2011;27(4):578–9.
Salmela L, Rivals E. LoRDEC: accurate and efficient long read error correction. Bioinformatics. 2014;30(24):3506–14.
Boetzer M, Pirovano W. SSPACE-LongRead: scaffolding bacterial draft genomes using long read sequence information. BMC Bioinformatics. 2014;15(1):211.
Simao FA, Waterhouse RM, Ioannidis P, Kriventseva EV, Zdobnov EM. BUSCO: assessing genome assembly and annotation completeness with single-copy orthologs. Bioinformatics. 2015;31(19):3210–2.
Zimin AV, Delcher AL, Florea L, Kelley DR, Schatz MC, Puiu D, Hanrahan F, Pertea G, Van Tassell CP, Sonstegard TS, et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10(4):R42.
Canavez FC, Luche DD, Stothard P, Leite KR, Sousa-Canavez JM, Plastow G, Meidanis J, Souza MA, Feijao P, Moore SS, et al. Genome sequence and assembly of Bos indicus. J Hered. 2012;103(3):342–8.
Tarailo-Graovac M, Chen N: Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 2009:4.10. 11–14.10. 14.
Bao W, Kojima KK, Kohany O. Repbase update, a database of repetitive elements in eukaryotic genomes. Mob DNA. 2015;6(1):11.
Huerta-Cepas J, Forslund K, Coelho LP, Szklarczyk D, Jensen LJ, von Mering C, Bork P. Fast Genome-wide functional annotation through Orthology assignment by eggNOG-mapper. Mol Biol Evol. 2017;34(8):2115–22.
Zdobnov EM, Apweiler R. InterProScan–an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17(9):847–8.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology Consortium. Nat Genet. 2000;25(1):25–9.
Kanehisa M, Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28(1):27–30.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M: KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res 2007, 35(suppl_2):W182-W185.
Prakash B, Dhali A, Rathore SS, Das KC, Walling I, Vupru K, Mech A, Baruah KK, Rajkhowa C. Chemical composition and nutritional evaluation of various foliages consumed by mithun (Bos frontalis). Anim Feed Sci Technol. 2009;150(3–4):223–9.
Mondal M, Baruah K, Rajkhowa C. Mithun: an animal of Indian pride. Lives Res Rural Deve. 2014;26.
** D, Liu Q, Guo J, Yu H, Yang Y, He Y, Mao H, Gou X, Deng W. Genetic variability of the coding region for the prion protein gene (PRNP) in gayal (Bos frontalis). Mol Biol Rep. 2012;39(2):2011–20.
Ross MG, Russ C, Costello M, Hollinger A, Lennon NJ, Hegarty R, Nusbaum C, Jaffe DB. Characterizing and measuring bias in sequence data. Genome Biol. 2013;14(5):R51.
Ikegami T, Inatsugi T, Kojima I, Umemura M, Hagiwara H, Machida M, Asai K. Hybrid De Novo Genome Assembly Using MiSeq and SOLiD Short Read Data. PLoS One. 2015;10(4):e0126289.
Miller JR, Zhou P, Mudge J, Gurtowski J, Lee H, Ramaraj T, Walenz BP, Liu J, Stupar RM, Denny R, et al. Hybrid assembly with long and short reads improves discovery of gene family expansions. BMC Genomics. 2017;18(1):541.
English AC, Richards S, Han Y, Wang M, Vee V, Qu J, Qin X, Muzny DM, Reid JG, Worley KC, et al. Mind the gap: upgrading genomes with Pacific Biosciences RS long-read sequencing technology. PLoS One. 2012;7(11):e47768.
Mostovoy Y, Levy-Sakin M, Lam J, Lam ET, Hastie AR, Marks P, Lee J, Chu C, Lin C, Dzakula Z, et al. A hybrid approach for de novo human genome sequence assembly and phasing. Nat Methods. 2016;13(7):587–90.
Putnam NH, O'Connell BL, Stites JC, Rice BJ, Blanchette M, Calef R, Troll CJ, Fields A, Hartley PD, Sugnet CW, et al. Chromosome-scale shotgun assembly using an in vitro method for long-range linkage. Genome Res. 2016;26(3):342–50.
Larsen PA, Harris RA, Liu Y, Murali SC, Campbell CR, Brown AD, Sullivan BA, Shelton J, Brown SJ, Raveendran M, et al. Hybrid de novo genome assembly and centromere characterization of the gray mouse lemur (Microcebus murinus). BMC Biol. 2017;15(1):110.
Sohn J-i, Nam K, Kim J-M, Lim D, Lee K-T, Do YJ, Cho CY, Kim N, Nam J-W, Chae H-H: Whole genome hybrid assembly and protein-coding gene annotation of the entirely black native Korean chicken breed Yeonsan Ogye. bioRxiv 2017:224311.
Gordon D, Huddleston J, Chaisson MJ, Hill CM, Kronenberg ZN, Munson KM, Malig M, Raja A, Fiddes I, Hillier LW, et al. Long-read sequence assembly of the gorilla genome. Sci. 2016;352(6281):aae0344.
Bashir A, Klammer AA, Robins WP, Chin C-S, Webster D, Paxinos E, Hsu D, Ashby M, Wang S, Peluso P. A hybrid approach for the automated finishing of bacterial genomes. Nat Biotechnol. 2012;30(7):701.
Koren S, Harhay GP, Smith TP, Bono JL, Harhay DM, McVey SD, Radune D, Bergman NH, Phillippy AM. Reducing assembly complexity of microbial genomes with single-molecule sequencing. Genome Biol. 2013;14(9):R101.
Pendleton M, Sebra R, Pang AW, Ummat A, Franzen O, Rausch T, Stutz AM, Stedman W, Anantharaman T, Hastie A, et al. Assembly and diploid architecture of an individual human genome via single-molecule technologies. Nat Methods. 2015;12(8):780–6.
Mukherjee S, Mukherjee A, Jasrotia RS, Jaiswal S, Iquebal MA, Longkumer I, Mech M, Vüpru K, Khate K, Rajkhowa C, et al. Muscle transcriptome signature and gene regulatory network analysis in two divergent lines of a hilly bovine species Mithun (Bos frontalis). Genomics. 2019.
Walker EP: Mammals of the world, vol. III. Hopkins, Baltimore 1968.
Ma G, Chang H, Li S, Chen H, Ji D, Geng R, Chang C, Li Y. Phylogenetic relationships and status quo of colonies for gayal based on analysis of cytochrome B gene partial sequences. J Genet Genomics. 2007;34(5):413–9.
Verkaar EL, Nijman IJ, Beeke M, Hanekamp E, Lenstra JA. maternal and paternal lineages in cross-breeding bovine species. Has wisent a hybrid origin. Mol Biol Evol. 2004;21(7):1165–70.
Nijman IJ, van Boxtel DCJ, van Cann LM, Marnoch Y, Cuppen E, Lenstra JA. Phylogeny of Y chromosomes from bovine species. Cladistics. 2008;24(5):723–6.
Hassanin A, Ropiquet A. Molecular phylogeny of the tribe Bovini (Bovidae, Bovinae) and the taxonomic status of the Kouprey, Bos sauveli Urbain 1937. Mol Phylogenet Evol. 2004;33(3):896–907.
Chi J, Fu B, Nie W, Wang J, Graphodatsky AS, Yang F. New insights into the karyotypic relationships of Chinese muntjac (Muntiacus reevesi), forest musk deer (Moschus berezovskii) and gayal (Bos frontalis). Cytogenet Genome Res. 2005;108(4):310–6.
Gallagher D Jr, Womack J. Chromosome conservation in the Bovidae. J Hered. 1992;83(4):287–98.
Kilkenny C, Browne WJ, Cuthill IC, Emerson M, Altman DG. Improving bioscience research reporting: the ARRIVE guidelines for reporting animal research. PLoS Biol. 2010;8:e1000412.
Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30(15):2114–20.
LoRDEC: a hybrid error correction program for long, PacBio reads. Assessed online. http://atgc.lirmm.fr/lordec/.
Worley KC: Improving Genomes Using Long Reads and PBJelly 2. In: Plant and Animal Genome XXII Conference: 2014: Plant and Animal Genome; 2014.
Gurevich A, Saveliev V, Vyahhi N, Tesler G. QUAST: quality assessment tool for genome assemblies. Bioinformatics. 2013;29(8):1072–5.
Li H: Aligning sequence reads, clone sequences and assembly contigs with BWA-MEM. ar** and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27(21):2987–93.
Waterhouse RM, Seppey M, Simão FA, Manni M, Ioannidis P, Klioutchnikov G, Kriventseva EV, and Zdobnov EM. BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol Biol Evol, published online Dec 6, 2017. https://doi.org/10.1093/molbev/msx319.
Marcais G, Kingsford C. A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics. 2011;27(6):764–70.
Guo LT, Wang SL, Wu QJ, Zhou XG, **e W, Zhang YJ. Flow cytometry and K-mer analysis estimates of the genome sizes of Bemisia tabaci B and Q (Hemiptera: Aleyrodidae). Front Physiol. 2015;6:144.
Haas BJ, Papanicolaou A, Yassour M, Grabherr M, Blood PD, Bowden J, Couger MB, Eccles D, Li B, Lieber M, et al. De novo transcript sequence reconstruction from RNA-seq using the trinity platform for reference generation and analysis. Nat Protoc. 2013;8(8):1494–512.
Kim D, Pertea G, Trapnell C, Pimentel H, Kelley R, Salzberg SL. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14(4):R36.
Trapnell C, Roberts A, Goff L, Pertea G, Kim D, Kelley DR, Pimentel H, Salzberg SL, Rinn JL, Pachter L. Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and cufflinks. Nat Protoc. 2012;7(3):562–78.
Slater GS, Birney E. Automated generation of heuristics for biological sequence comparison. BMC Bioinformatics. 2005;6(1):31.
Consortium U. The universal protein resource (UniProt). Nucleic Acids Res. 2008;36(suppl 1):D190–5.
Haas BJ, Delcher AL, Mount SM, Wortman JR, Smith RK, Hannick LI, Maiti R, Ronning CM, Rusch DB, Town CD, et al. Improving the Arabidopsis genome annotation using maximal transcript alignment assemblies. Nucleic Acids Res. 2003;31(19):5654–66.
Stanke M, Keller O, Gunduz I, Hayes A, Waack S, Morgenstern B. AUGUSTUS: ab initio prediction of alternative transcripts. Nucleic Acids Res. 2006;34(Web Server issue):W435–9.
Haas BJ, Salzberg SL, Zhu W, Pertea M, Allen JE, Orvis J, White O, Buell CR, Wortman JR. Automated eukaryotic gene structure annotation using EVidenceModeler and the program to assemble spliced alignments. Genome Biol. 2008;9(1):R7.
Bateman A, Coin L, Durbin R, Finn RD, Hollich V, Griffiths-Jones S, Khanna A, Marshall M, Moxon S, Sonnhammer EL, et al. The Pfam protein families database. Nucleic Acids Res. 2004;32(Database issue):D138–41.
Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003;13(9):2129–41.
Attwood TK, Blythe MJ, Flower DR, Gaulton A, Mabey JE, Maudling N, McGregor L, Mitchell AL, Moulton G, Paine K, et al. PRINTS and PRINTS-S shed light on protein ancestry. Nucleic Acids Res. 2002;30(1):239–41.
Yeats C, Maibaum M, Marsden R, Dibley M, Lee D, Addou S, Orengo CA. Gene3D: modelling protein structure, function and evolution. Nucleic Acids Res. 2006;34(Database issue):D281–4.
Wilson D, Pethica R, Zhou Y, Talbot C, Vogel C, Madera M. SUPERFAMILY–comparative genomics, datamining and sophisticated visualization. Nucleic Acids Res. 2009;37:D380–6.
Moriya Y, Itoh M, Okuda S, Yoshizawa AC, Kanehisa M. KAAS: an automatic genome annotation and pathway reconstruction server. Nucleic Acids Res. 2007;35(Web Server issue):W182–5.
Okuda S, Yamada T, Hamajima M, Itoh M, Katayama T, Bork P, Goto S, Kanehisa M. KEGG Atlas map** for global analysis of metabolic pathways. Nucleic Acids Res. 2008;36(Web Server issue):W423–6.
Harris RS. Improved pairwise alignment of genomic DNA: the Pennsylvania State University; 2007.
Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc Natl Acad Sci U S A. 2003;100(20):11484–9.
Acknowledgements
The authors would like to thank the Director of the Institute and Indian Council of Agricultural Research for funding the Institutional Research Project for this study. Financial assistance in the form of DBT Overseas Associateship for the first author is gratefully acknowledged. IL and MM were supported by a DST and DBT funded grant, Govt. Of India, respectively. Assistance of DBT Biotech Hub in carrying out a part of wet lab work is thankfully acknowledged.
Funding
This work was carried out under an Institutional research project funded by ICAR-NRC on Mithun, Nagaland, India. Financial support in the form of DBT Overseas Associateship for the first author played a significant role to carry out some of the bioinformatic analyses and their interpretations at Aarhus University, Denmark.
Author information
Authors and Affiliations
Contributions
SM, AM and CR conceived the study. SM, ZC and AM run analysis and wrote the manuscript consulting other authors. IL, MM, KV, KK collected samples and run wet lab work. ABM, BG, MS, and GS critically read and revised the manuscript. All authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Ethics approval and consent to participate
All the animal experiments and sample collections procedures were carried out by qualified Veterinarians of the Institute. The ethics approval for animal experimentation was obtained from the Institute Ethics Approval Committee under the mandatory regulations of CPCSEA (Committee for the Purpose of Control and Supervision on Experiments on Animals) guidelines, prescribed by the Indian Council of Agricultural Research (ICAR), Ministry of Agriculture and Farmers Welfare, Government of India. Since no specimen was used, no permissions were necessary to collect the specimens in our present study.
Consent for publication
Not Applicable
Competing interests
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Additional files
Additional file 1:
NC3Rs ARRIVE Guidelines Checklist. (PDF 160 kb)
Additional file 2:
Table S1. Genes found in mithun genome with at least one domain hit. (XLSX 2390 kb)
Additional file 3:
Table S2. Genes found in mithun genome with GO analysis. (XLSX 5227 kb)
Additional file 4:
Table S3. Genes found in mithun genome with KEGG entries. (XLSX 510 kb)
Rights and permissions
Open Access This article is distributed under the terms of the Creative Commons Attribution 4.0 International License (http://creativecommons.org/licenses/by/4.0/), which permits unrestricted use, distribution, and reproduction in any medium, provided you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The Creative Commons Public Domain Dedication waiver (http://creativecommons.org/publicdomain/zero/1.0/) applies to the data made available in this article, unless otherwise stated.
About this article
Cite this article
Mukherjee, S., Cai, Z., Mukherjee, A. et al. Whole genome sequence and de novo assembly revealed genomic architecture of Indian Mithun (Bos frontalis). BMC Genomics 20, 617 (2019). https://doi.org/10.1186/s12864-019-5980-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1186/s12864-019-5980-y