Abstract
A minimally redundant set of 15,000 mouse cDNAs derived from early and mid-gestation has been used to analyze global patterns of gene expression in the placenta and embryo.
Similar content being viewed by others
Significance and context
The analysis of gene expression during early embryonic development is difficult in humans, and so many scientists instead make use of 'model' organisms such as the mouse. Tanaka et al. have constructed a cDNA array of genes expressed at this time, which can be used as an exploratory tool to investigate gene expression profiles at this stage of development. The authors used the array to identify genes that have elevated expression in the placenta compared with the embryo. Profiling of placenta-specific genes should aid the determination of the mechanisms of placental development and maintenance. An important aspect of this study is the attention paid to statistical significance, with all experiments performed in triplicate.
Key results
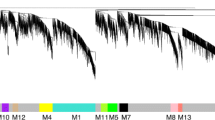
Just over 52,000 mouse expressed sequence tags (ESTs), mainly from pre- and peri-implantation embryos were clustered into a minimally redundant set of 15,000 clones which were then used to construct a cDNA microarray. The number of genes found to be highly expressed in or specific to the placenta in this study was fivefold greater than in previous studies. One of the placenta-specific genes encoded a novel member of the placental growth-related hormone family.
Links
Details of the National Institute of Aging mouse cDNA project, the ESTs used in construction of the 15K array, and all hybridization data can be found through the NIA/NIH mouse genomics homepage.
Conclusions
The arrayed cDNAs will be invaluable in studying early murine development, and will also find application in analysis of reactivation of embryonic genes during the process of aging.
Reporter's comments
This study has produced one of the first large collections of non-redundant, sequence-validated mouse cDNAs suitable for microarray construction and has demonstrated the power of expression profiling to identify novel, placenta-specific genes.
References
Tanaka TS, Jaradat SA, Lim MK, Kargul GJ, Wang X, Grahovac MJ, Pantano S, Sano Y, Piao Y, Nagaraja R, et al: Genome-wide expression profiling of mid-gestation placenta and embryo using a 15K mouse developmental cDNA microarray. Proc Natl Acad Sci U S A. 2000, 97: 9127-9132. 0027-8424
Rights and permissions
About this article
Cite this article
Denny, P. Mouse placental gene expression . Genome Biol 1, reports0069 (2000). https://doi.org/10.1186/gb-2000-1-3-reports0069
Received:
Published:
DOI: https://doi.org/10.1186/gb-2000-1-3-reports0069