Abstract
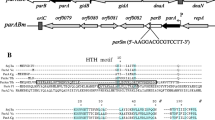
Stable inheritance of bacterial chromosomes and low copy number plasmids is ensured by accurate partitioning of replicated molecules between the daughter cells at division. Partitioning of the prophage of the temperate bacteriophage N15, which exists as a linear plasmid molecule with covalently closed ends, depends on the sop locus, comprising genes sopA and sopB, as well as four centromere sites in different regions of the N15 genome essential for replication and the control of lysogeny. We found that binding of SopB to the centromere could silence centromere-proximal promoters, presumably due to subsequent polymerization of SopB along the DNA. Close to the IR4 centromere site we identified a promoter, P59, which was able to drive the expression of phage late genes encoding structural proteins of virion. We found that, following binding to IR4, the N15 Sop proteins could induce repression of this promoter. The repression depended on SopB and was enhanced in the presence of SopA. Sop-dependent silencing of centromere-proximal promoters may control gene expression in phage N15, particularly preventing undesired expression of late genes in the N15 prophage. Thus, the phage N15 sop system not only ensures plasmid partitioning but is also involved in the genetic network controlling prophage replication and the maintenance of lysogeny.
Similar content being viewed by others
References
Austin S., Abeles A. 1983. Partition of unit-copy miniplasmids to daughter cells: 2. The partition region of miniplasmid P1 encodes an essential protein and a centromere-like site at which it acts. J. Mol. Biol. 169, 373–387.
Ogura T., Hiraga S. 1983. Partition mechanism of F plasmid: Two plasmid gene-encoded products and a cis-acting region are involved in partition. Cell. 32, 351–360.
Gerdes K., Moller-Jensen J., Jensen R.B. 2000. Plasmid and chromosome partitioning: Surprises from phylogeny. Mol. Microbiol. 37, 455–466.
Mori H., Mori Y., Ichinose C., Niki H., Ogura T., Kato A., Hiraga S. 1989. Purification and characterization of SopA and SopB proteins essential for F plasmid partitioning. J. Biol. Chem. 264, 15535–15541.
Friedman S.A., Austin S.J. 1988. The P1 plasmid-partition system synthesizes two essential proteins from an autoregulated operon. Plasmid. 19, 103–112.
Hayakawa Y., Murotsu T., Matsubara K. 1985. Mini-F protein that binds to a unique region for partition of mini-F plasmid DNA. J. Bacteriol. 163, 349–354.
Yates P., Lane D., Biek D.P. 1999. The F plasmid centromere, sopC, is required for full repression of the sopAB operon. J. Mol. Biol. 290, 627–638.
Ravin N.V., Dorokhov B.D., D. Lane. 2004. Structural organization and control of expression of the sop operon of linear plasmid prophage N15. Mol. Biol. 38, 247–252.
Hao J.J., Yarmolinsky M. 2002. Effects of the P1 plasmid centromere on expression of P1 partition genes. Mol. Boil. (Moscow), 38, 297–302.
Gordon G.S., Sitnikov D., Webb C.D., Teleman A., Straight A., Losick R., Murray A.W., Wright A. 1997. Chromosome and low copy plasmid segregation in E. coli: Visual evidence for distinct mechanisms. Cell. 90, 1113–1121.
Moller-Jensen J., Jensen R.B., Lowe J., Gerdes K. 2002. Prokaryotic DNA segregation by an actin-like filament. EMBO J. 21, 3119–3127.
Ravin V.K., Shulga M.G. 1970. Evidence for extrachromosomal location of prophage N15. Virology. 40, 800–807.
Svarchavskii A.N., Rybchin V.N. 1984. Physical map** of plasmid N15 DNA. Mol. Genet. Mikrobiol. Virusol. 10, 16–22.
Ravin N.V. 2003. Mechanisms of replication and telomere resolution of the linear plasmid prophage N15. FEMS Microbiol. Lett. 221, 1–6.
Barbour A.G., Garon C.F. 1987. Linear plasmids of the Borrelia burgdorferi have covalently closed ends. Science. 237, 409–411.
Allardet-Servent A., Michaux-Charachon S., Jumas-Bilak E., Karayan L., Ramuz M. 1993. Presence of one linear and one circular chromosome in the Agrobacterium tumefaciens C58 genome. J. Bacteriol. 175, 7869–7874.
Hertwig S., Klein I., Lurz R., Lanka E., Appel B. 2003. PY54, a linear plasmid prophage of Yersinia enterocolitica with covalently closed ends. Mol. Microbiol. 48, 989–1003.
Casjens S.R., Gilcrease E.B., Huang W.M., Bunny K.L., Pedulla M.L., Ford M.E., Hourtz J.M., Hatfull G.F., Hendrix R.W. 2004. The pKO2 linear plasmid prophage of Klebsiella oxytoca. J. Bacteriol. 186, 1818–1832.
Ravin N., Lane D. 1999. Partition of the linear plasmid N15: Interactions of N15 partition functions with the sop locus of the F plasmid. J. Bacteriol. 181, 6898–6906.
Grigoriev P.S., Lobocka M.B. 2001. Determinants of segregational stability of the linear plasmid-prophage N15 of Escherichia coli. Mol. Microbiol. 42, 355–368.
Kostelidou K., Thomas C.M. 2000. The hierarchy of KorB binding at its 12 binding sites on the broad-host-range plasmid RK2 and modulation of this binding by IncC1 protein. J. Mol. Biol. 295, 411–422.
Chiu C.M., Manzoor S.E., Batt S.M., Muntaha S., Bingle L.E., Thomas C.M. 2008. Distribution of the partitioning protein KorB on the genome of IncP-1 plasmid RK2. Plasmid. 59, 163–175.
Ravin N.V., Rech J., Lane D. 2008. Extended function of plasmid partition genes: Sop system of linear phage-plasmid N15 facilitates late gene expression. J. Bacteriol. 190, 3538–3545.
Rodionov O., Lobocka M., Yarmolinsky M. 1999. Silencing of genes flanking the P1 plasmid centromere. Science. 283, 546–549.
Casadaban M.J., Cohen S.N. 1980. Analysis of gene control signals by DNA fusion and cloning in Escherichia coli. J. Mol. Biol. 138, 179–207.
Grant S.G., Jessee J., Bloom F.R., Hanahan D. 1990. Differential plasmid rescue from transgenic mouse DNAs into Escherichia coli methylation-restriction mutants. Proc. Natl. Acad. Sci. USA. 87, 4645–4649.
Ravin V., Ravin N., Casjens S., Ford M., Hatfull G., Hendrix R. 2000. Genomic sequence and analysis of the atypical bacteriophage N15. J. Mol. Biol. 299, 53–73.
Ravin N.V., Rech J., Lane D. 2003. Map** of functional domains in F plasmid partition proteins reveals a bipartite SopB-recognition domain in SopA. J. Mol. Biol. 329, 875–889.
Lemonnier M., Lane D. 1998. Expression of the second lysine decarboxylase gene of Escherichia coli. Microbiology. 144, 751–760.
Simons R.W., Houman F., Kleckner N. 1987. Improved single and multicopy lac-based cloning vectors for protein and operon fusions. Gene. 53, 85–96.
Miller J.H. 1972. Experiments in Molecular Genetics. Cold Spring Harbor, NY: Cold Spring Harbor Lab. Press.
Lynch A.S., Wang J.C. 1995. SopB protein-mediated silencing of genes linked to the sopC locus of Escherichia coli F plasmid. Proc. Natl. Acad. Sci. USA. 92, 1896–1900.
Rodionov O.M., Yarmolinsky M. 2004. Plasmid partitioning and the spreading of P1 partition protein ParB. Mol. Microbiol. 52, 1215–1223.
Author information
Authors and Affiliations
Corresponding author
Additional information
Original Russian Text © A.V. Mardanov, D. Lane, N.V. Ravin, 2010, published in Molekulyarnaya Biologiya, 2010, Vol. 44, No. 2, pp. 294–300.
Rights and permissions
About this article
Cite this article
Mardanov, A.V., Lane, D. & Ravin, N.V. Sop proteins can cause transcriptional silencing of genes located close to the centromere sites of linear plasmid N15. Mol Biol 44, 262–267 (2010). https://doi.org/10.1134/S0026893310020111
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1134/S0026893310020111