Abstract
Iris germanica L. is a perennial herbaceous plant that has been widely cultivated worldwide and is popular for its elegant and vibrantly colorful flowers. Selection of appropriate reference genes is the prerequisite for accurate normalization of target gene expression by quantitative real-time PCR. However, to date, the most suitable reference genes for flowering stages have not been elucidated in I. germanica. In this study, eight candidate reference genes were examined for the normalization of RT-qPCR in three I. germanica cultivars, and their stability were evaluated by four different algorithms (GeNorm, NormFinder, BestKeeper, and Ref-finder). The results revealed that IgUBC and IgGAPDH were the most stable reference genes in ‘00246’ and ‘Elizabeth’, and IgTUB and IgUBC showed stable expression in ‘2010200’. IgUBC and IgGAPDH were the most stable in all samples, while IgUBQ showed the least stability. Finally, to validate the reliability of the selected reference genes, the expression patterns of IgFT (Flowering Locus T gene) was analyzed and emphasized the importance of appropriate reference gene selection. This work presented the first systematic study of reference genes selection during flower bud development and provided guidance to research of the molecular mechanisms of flowering stages in I. germanica.
Similar content being viewed by others
Introduction
Quantitative real-time PCR (RT-qPCR) is a reliable and widely used technique to quantify target gene expression patterns in various fields of biological research, due to its high sensitivity, accuracy and reproducibility1,2,3. However, the accuracy of RT-qPCR is influenced by various factors, including the quantity of mRNA templates, enzymatic efficiency in cDNA synthesis and PCR amplification4. Thus, to avoid bias, it is necessary to select reliable reference genes that are steadily expressed under different experimental conditions before determining the expression pattern of a target gene by RT-qPCR.
Traditional reference genes, such as elongation factor 1 alpha (EF1α), glyceraldehyde-3-phosphate dehydrogenase (GAPDH), actin (ACT) and ubiquitin (UBQ), are mostly involved in intermediary metabolism or other basic cellular processes5,12, have been developed for the evaluation of potential reference gene(s) in different experimental systems.
Flowers of higher plants are reproductive organs that are widely studied and are important ornamental characteristics of ornamental plants. A number of reference genes have been reported for different flowering stages of various ornamental plants species, such as chrysanthemum13, petunia14, azalea15 and tree peony16, but little information is available concerning reference genes for Iris. Iris germanica L., which is often referred to as Pogon iris, is one of the most important ornamental species in the Iris genus17, the posture of the flower is peculiar and the flowers are rich and have high ornamental and economic value. However, the spring-time flowering habit and short duration of flowering of I. germanica hinder its year-round supply and economic benefits. To enable additional detailed and in-depth studies of the expression level of key genes involved in flowering, it is necessary to identify the stability of candidate reference genes at various flowering stages in I. germanica. Recently, studies have been shown that ACT11 performed well in different tissues but poorly in different flower development stages in I. germanica cultivar ‘Huang**jia’. Furthermore, TUA performed best in different flower development stages but was the worst in different tissues18. Nevertheless, limited information is available concerning reference genes for flowering stages in I. germanica.
In the present study, the expression stability of eight candidate reference genes, IgEF1α, IgGAPDH, IgACT6, IgUBQ, IgUBC (ubiquitin-protein ligase), IgEF1β (elongation factor 1β), IgPGK (phosphoglycerate kinase), IgTUB (beta-tubulin), was validated by RT-qPCR during flower development in different cultivars of I. germanica. Four statistical algorithms, GeNorm10, BestKeeper12, NormFinder11, and Ref-finder (http://www.leonxie.com/referencegene.php) were used to evaluate the most suitable reference genes needed for normalization. To verify the usefulness of the selected reference genes, the relative expression levels of IgFT, a putative homolog of Flowering Locus T (FT) gene in Arabidopsis, was analyzed during flower bud development and in different genotypes of I. germanica. This is the first report about the selection of reference genes during flower bud development and will benefit future studies on gene expression of flowering stages in I. germanica and other related species.
Results
Assessment of primer specificity and amplification efficiency of reference genes
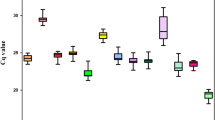
The gene names, primer sequences and amplicon length characteristics of the 8 reference genes are summarized in Table 1. The primer specificities were verified by agarose gel electrophoresis, only a specific product of the expected size was observed, and no primer dimers were detected (Supplementary Fig. S1). In addition, only a single peak was found in the melting curves of the amplified products of all genes, indicating that no primer dimers were generated (Supplementary Fig. S2). The amplification efficiency (E) of each reference gene varied from 98.47% for IgGAPDH to 101.74% for IgUBQ, and the correlation coefficients (R2) ranged from 0.9984 to 0.9998 (Table 1). The amplicon size ranged from 107 for IgACT6 to 295 bp for IgUBQ.
Expression levels and profiles of reference genes
The quantification cycle (Cq) values of eight reference genes were assayed by RT-qPCR analysis, with lower Cq values representing relatively higher mRNA transcript levels. Cq values for each of the 8 candidate reference genes in three different genotypes of I. germanica are listed in supplementary Table S1. The average Cq values of these reference genes ranged from 19.83 to 27.61 for the highest and lowest expression levels, respectively, across all samples (Fig. 1). In addition, the standard deviations (SD) of Cq values were used to determine reference gene stability levels. Genes with relatively high SD of Cq values showed more variable expression than did those with lower SD. IgGAPDH showed the least variation in gene expression (23.69 ± 0.36) and presented the lowest SD, while IgEF1β showed the most variable levels of expression (23.16 ± 1.38).
Expression stability of reference genes
To further select the most appropriate reference gene for RT-qPCR-based analysis in the investigation of flower development across three different I. germanica cultivars, four software programs, GeNorm, NormFinder, BestKeeper and RefFinder were used to analyze the expression stability of each reference gene.
GeNorm analysis
GeNorm program was used to evaluate the expression stability of the 8 candidate reference genes by calculating average expression stability (M) values based on the average pairwise variation among all the tested genes. According to GeNorm algorithm, stably expressed genes had M values below 1.5, and a relatively low M value indicates a relatively stable expression10. In this study, all of the tested genes showed high expression stability, with M-values of < 1.5, indicating that they all conformed to basic requirements for function as reference genes. IgUBC and IgGAPDH were the most stable reference genes in both ‘00246’ and ‘Elizabeth’, while IgTUB and IgGAPDH were identified as the most stable in ‘2010200’. In terms of the total samples tested, IgUBC and IgGAPDH were recommended as the most stable reference genes. In contrast, IgUBQ with the highest M value was identified as the least stable reference gene in all of the samples (Fig. 2).
The optimal number of reference genes was also measured by determining the pairwise variation between sequentially ranked genes (Vn/Vn + 1) based on the GeNorm algorithm (Fig. 3). Generally speaking, a cutoff of 0.15 (Vn value) has been recommended as the threshold to determine the optimal number of reference genes10. Our results reveal that the V2/3 values of the ‘00246’, ‘2010200’ and ‘Elizabeth’ samples were lower than 0.15 (Fig. 3), suggesting that two reference genes were sufficient for accurate normalization. However, the value of 0.15 should not be a fixed threshold, and higher cutoff values of Vn/n + 1 have been shown in several reports19,20. Our data showed small variation between V2/3 and V3/4 across all the samples, suggesting that two reference genes were sufficient for normalization (Fig. 3), which was similar to results in bermudagrass21 and Kentucky bluegrass22.
NormFinder analysis
NormFinder is used to determine the stability value of reference genes, based on inter- and intragroup variance in different sample groups11. The stability value is then calculated, with a relatively low stability value meaning that the gene is relatively stable. Reference gene stability values were calculated by NormFinder, as shown in Table 2. IgUBC and IgGAPDH were the two most stable genes among the total group, while IgUBQ was the least stable. The top two most stably expressed genes were IgUBC and IgGAPDH in both ‘00246’ and ‘2010200’, and IgUBC and IgTUB in ‘Elizabeth’. The ranking order generated by this method was slightly different from that of GeNorm.
BestKeeper analysis
BestKeeper evaluates the stability of reference genes based on the standard deviation (SD) and coefficients of variation (CV) of Cq values, with relatively low SD and CV representing relatively high stability12. The results of BestKeeper analysis are listed in Table 3. IgGAPDH and IgUBC were recommended as the most stable genes in ‘00246’, ‘Elizabeth’ and across all the samples, which was similar to the results from the GeNorm and NormFinder analysis. In ‘2010200’ samples, IgTUB and IgACT6 were detected as the most stable genes via BestKeeper analysis, whereas IgACT6 was ranked fourth by GeNorm and sixth by NormFinder.
RefFinder analysis
RefFinder (http://www.leonxie.com/referencegene.php) was used to obtain the comprehensive rankings of reference genes by integrating three common analysis programs: GeNorm, NormFinder and BestKeeper13,27. The putative homologs of 8 reference genes were identified from the transcriptomic data sequences of Iris lactea var. chinensis49. All the candidate reference genes were cloned in Stage 4 samples and confirmed through sequencing. Primers were designed using Primer 5.0 software (Premier Biosoft International) with melting temperatures (TM) of 55–65 ℃, primer lengths of 18–25 bp and amplicon lengths of 107–295 bp (Table 1). The performance of the primers was tested by qPCR and the specificity of the primer amplicons was further verified by 2% (w/v) gel electrophoresis.
Quantitative real-time PCR (RT-qPCR)
qPCR was performed using a Mastercycler ep realplex 2S device (Eppendorf, Germany) in conjunction with SYBR Premix Ex Taq II (TaKaRa, Dalian, China). Reactions were performed in a total volume of 20 μL containing 5 μL of diluted cDNA, 0.6 μL of each of forward and reverse primer (10 μM), 10 μL of 2 × SYBR Premix and 3.8 μL of ddH2O. The amplification program comprised an initial denaturation step (95℃ for 2 min), followed by 40 cycles of 95℃ for 5 s, 60℃ for 30 s, and 72 ℃ for 30 s, and a melting curve protocol (60–95 ℃ with a temperature increment of 0.5 ℃ s−1). Each RT-qPCR was performed for three biological and three technical replicates, and negative controls were included for each primer pair. Amplification efficiency (E) and correlation coefficient (R2) values were obtained from standard curves generated using a tenfold diluted cDNA series, the starting quantity of cDNA was 500 ng/μL50.
Data analysis
The stability of the eight candidate reference genes was assessed using GeNorm10, NormFinder software11, BestKeeper12, and Ref-finder (http://www.leonxie.com/referencegene.php). For GeNorm and NormFinder analysis, quantification cycle (Cq) values were transformed into relative expression levels using the formula: 2−∆Cq, in which ∆Cq = each corresponding Cq value-the minimum Cq value16. The expression stability measurement (M) was determined by the GeNorm program based on the average pairwise variations (V) among all the other reference genes. NormFinder program estimates intra- and intergroup variations, and the lowest stability is ranked the highest. Calculations of the BestKeeper program are calculated based on the coefficients of variation (CV) and the standard deviations (SD) of the Cq values, and the lowest CV and SD were used as detection indexes for the most-stable reference genes. Ref-finder is an online tool that integrates the results of the currently available major computational programs, including GeNorm (M values), NormFinder (stability values), BestKeeper (CV and SD), and ∆Cq values.
Validation of selected reference genes
To validate the influence of the choice of different reference genes on the final normalized outcome, the relative expression levels of IgFT which plays an important role in promoting flowering51 in three cultivars were analyzed using individual stably expressed or unstably expressed genes or a combination of stable reference genes, as determined by GeNorm48. The primers used for IgFT are presented in Table 1. The fold change of gene expression was calculated using the 2−∆∆Ct method24.
Conclusions
This research provided the first systematic analysis for the selection of stable reference genes as the internal control in RT-qPCR analysis in different flowering stages and different genotypes of I. germanica cultivars. Analysis using GeNorm, NormFinder, BestKeeper, and Ref-finder revealed that IgUBC, IgGAPDH, and IgTUB could be considered as appropriate reference genes for gene expression analysis in future molecular researches that aim to understand the mechanisms of flowering stages in I. germanica.
References
VanGuilder, H. D., Vrana, K. E. & Freeman, W. M. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques 44, 619–626. https://doi.org/10.2144/000112776 (2008).
Bustin, S. A. et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 55, 611–622. https://doi.org/10.1373/clinchem.2008.112797 (2009).
Artico, S., Nardeli, S. M., Brilhante, O., Grossi-de-Sa, M. F. & Alves-Ferreira, M. Identification and evaluation of new reference genes in Gossypium hirsutum for accurate normalization of real-time quantitative RT-PCR data. BMC Plant Biol. 10, 49. https://doi.org/10.1186/1471-2229-10-49 (2010).
Derveaux, S., Vandesompele, J. & Hellemans, J. How to do successful gene expression analysis using real-time PCR. Methods 50, 227–230. https://doi.org/10.1016/j.ymeth.2009.11.001 (2010).
Kozera, B. & Rapacz, M. Reference genes in real-time PCR. J. Appl. Genet. 54, 391–406. https://doi.org/10.1007/s13353-013-0173-x (2013).
**ao, X. L. et al. Validation of suitable reference genes for gene expression analysis in the halophyte Salicornia europaea by real-time quantitative PCR. Front. Plant Sci. 5, 788. https://doi.org/10.3389/fpls.2014.00788 (2014).
Delporte, M., Legrand, G., Hilbert, J. L. & Gagneul, D. Selection and validation of reference genes for quantitative real-time PCR analysis of gene expression in Cichorium intybus. Front. Plant Sci. 6, 651. https://doi.org/10.3389/fpls.2015.00651 (2015).
Niu, X. P. et al. Selection of reliable reference genes for quantitative real-time PCR gene expression analysis in Jute (Corchorus capsularis) under stress treatments. Front. Plant Sci. 6, 848. https://doi.org/10.3389/fpls.2015.00848 (2015).
Zhang, Y. X. et al. Selection of suitable reference genes for quantitative real-time PCR gene expression analysis in Salix matsudana under different abiotic stresses. Sci. Rep. 7, 40290. https://doi.org/10.1038/srep40290 (2017).
Vandesompele, J. et al. Accurate normalization of real-time quantitative RT-PCR data by geometric averaging of multiple internal control genes. Genome Biol. https://doi.org/10.1186/gb-2002-3-7-research0034 (2002).
Andersen, C. L., Jensen, J. L. & Ørntoft, T. F. Normalization of real-time quantitative reverse transcription-PCR data: a model-based variance estimation approach to identify genes suited for normalization, applied to bladder and colon cancer data sets. Cancer Res. 64, 5245–5250. https://doi.org/10.1158/0008-5472.CAN-04-0496 (2004).
Pfaffl, M. W., Tichopad, A., Prgomet, C. & Neuvians, T. P. Determination of stable housekee** genes, differentially regulated target genes and sample integrity: bestkeeper-Excel-based tool using pair-wise correlations. Biotechnol. Lett. 26, 509–515. https://doi.org/10.1023/B:BILE.0000019559.84305.47 (2004).
Qi, S. et al. Reference gene selection for RT-qPCR analysis of flower development in Chrysanthemum morifolium and Chrysanthemum lavandulifolium. Front. Plant Sci. 7, 287. https://doi.org/10.3389/fpls.2016.00287 (2016).
Mallona, I., Lischewski, S., Weiss, J., Hause, B. & Egea-Cortines, M. Validation of reference genes for quantitative real-time PCR during leaf and flower development in Petunia hybrida. BMC Plant Biol. 10, 4–14. https://doi.org/10.1186/1471-2229-10-4 (2010).
Yi, S. J. et al. Selection of reliable reference genes for gene expression studies in Rhododendron micranthum Turcz. Sci. Hortic-Amsterdam 138, 128–133. https://doi.org/10.1016/j.scienta.2012.02.013 (2012).
Li, J., Han, J. G., Hu, Y. H. & Yang, J. Selection of reference genes for quantitative real-time PCR during flower development in tree peony (Paeonia suffruticosa Andr). Front. Plant Sci. 7, 516. https://doi.org/10.3389/fpls.2016.00516 (2016).
Linnegar, S. & Hewitt, J. Irises (Octopus Publishing Group, 2008).
Wang, Y. J. et al. Reference gene selection for qRT-PCR normalization in Iris germanica L. Phyton Int. J. Exp. Bot. 90, 277–290. https://doi.org/10.32604/phyton.2020.011545 (2020).
Silveira, É. D., Alves-Ferreira, M., Guimarães, L. A., da Silva, F. R. & Carneiro, V. T. Selection of reference genes for quantitative real-time PCR expression studies in the apomictic and sexual grass Brachiaria brizantha. BMC Plant Biol. 9, 84. https://doi.org/10.1186/1471-2229-9-84 (2009).
Marum, L., Miguel, A., Ricardo, C. P. & Miguel, C. Reference gene selection for quantitative real-time PCR normalization in Quercus suber. PLoS ONE 7, e35113. https://doi.org/10.1371/journal.pone.0035113 (2012).
Niu, K. J., Shi, Y. & Ma, H. L. Selection of candidate reference genes for gene expression analysis in Kentucky Bluegrass (Poa pratensis L.) under abiotic stress. Front. Plant Sci. 8, 193. https://doi.org/10.3389/fpls.2017.00193 (2017).
Chen, Y. et al. Selection and validation of reference genes for target gene analysis with quantitative RT-PCR in leaves and roots of bermudagrass under four different abiotic stresses. Physiol. Plant. 155, 138–148. https://doi.org/10.1111/ppl.12302 (2014).
**e, F. L., **ao, P., Chen, D. L., Xu, L. & Zhang, B. H. miRDeepFinder: a miRNA analysis tool for deep sequencing of plant small RNAs. Plant Mol. Biol. 80, 75–84. https://doi.org/10.1007/s11103-012-9885-2 (2012).
Huggett, J. F., Dheda, K., Bustin, S. A. & Zumla, A. Real-time RT-PCR normalisation; strategies and considerations. Genes Immun. 6(279–284), 2012. https://doi.org/10.1038/sj.gene.6364190 (2005).
Chen, Y. et al. Cold acclimation induces freezing tolerance via antioxidative enzymes, proline metabolism and gene expression changes in two chrysanthemum species. Mol. Biol. Rep. 41, 815–822. https://doi.org/10.1007/s11033-013-2921-8 (2014).
Chapman, J. R. & Waldenström, J. With reference to reference genes: a systematic review of endogenous controls in gene expression studies. PLoS ONE 10, e0141853. https://doi.org/10.1371/journal.pone.0141853 (2015).
Wang, M. & Lu, S. Validation of suitable reference genes for quantitative gene expression analysis in Panax ginseng. Front. Plant Sci. 6, 1259. https://doi.org/10.3389/fpls.2015.01259 (2016).
Galli, G. G. et al. YAP drives growth by controlling transcriptional pause release from dynamic enhancers. Mol. Cell 60, 328–337. https://doi.org/10.1016/j.molcel.2015.09.001 (2015).
He, Y., Yan, H., Hua, W., Huang, Y. & Wang, Z. Selection and validation of reference genes for quantitative real-time PCR in Gentiana macrophylla. Front. Plant Sci. 7, 945. https://doi.org/10.3389/fpls.2016.00945 (2016).
Tang, F. et al. Selection and validation of reference genes for quantitative expression analysis of miRNAs and mRNAs in Poplar. Plant Methods 15, 35. https://doi.org/10.1186/s13007-019-0420-1 (2019).
Shivhare, R. & Lata, C. Selection of suitable reference genes for assessing gene expression in pearl millet under different abiotic stresses and their combinations. Sci. Rep. 6, 23036. https://doi.org/10.1038/srep23036 (2016).
Kim, H. et al. RNA-Seq analysis of spatiotemporal gene expression patterns during fruit development revealed reference genes for transcript normalization in plums. Plant Mol. Biol. Rep. 33, 1634–1649. https://doi.org/10.1007/s11105-015-0860-3 (2015).
**ao, Z. et al. Selection of reliable reference genes for gene expression studies on Rhododendron molle G. Don. Front. Plant Sci. 7, 1547. https://doi.org/10.3389/fpls.2016.01547 (2016).
Wang, C. et al. Identification and validation of reference genes for RT-qPCR analysis in non-heading Chinese cabbage flowers. Front. Plant Sci. 7, 811. https://doi.org/10.3389/fpls.2016.00811 (2016).
Fu, J., Wang, Y., Huang, H., Zhang, C. & Dai, S. L. Reference gene selection for RT-qPCR analysis of Chrysanthemum lavandulifolium during its flowering stages. Mol. Breed. 31, 205–215. https://doi.org/10.1007/s11032-012-9784-x (2013).
Fulvio, F., Martinelli, T. & Paris, R. Selection and validation of reference genes for RT-qPCR normalization indifferent tissues of milk thistle (Silybum marianum, Gaert). Gene Gene https://doi.org/10.1016/j.gene.2020.145272 (2020).
Gu, C. S. et al. Validation of reference genes for RT-qPCR normalization in Iris. lacteavar. chinensis leaves under different experimental conditions. Sci. Hortic-Amsterdam 175, 144–149. https://doi.org/10.1016/j.scienta.2014.06.011 (2014).
Ma, L. L. et al. Selection and validation of reference genes for quantitative real-time PCR analysis in Iris bulleyana during flower color variation. J. Nucl. Agric. Sci. 33, 1707–1716 (2019).
Saddhe, A., Malvankar, M. & Kumar, K. Selection of reference genes for quantitative real-time PCR analysis in halophytic plant Rhizophora apiculate. PeerJ 6, e5226. https://doi.org/10.7717/peerj.5226 (2018).
Ma, R., Xu, S., Zhao, Y., **a, B. & Wang, R. Selection and validation of appropriate reference genes for quantitative real-time PCR analysis of gene expression in Lycoris aurea. Front. Plant Sci. 7, 536. https://doi.org/10.3389/fpls.2016.00536 (2016).
Fan, C. et al. Selection of reference genes for quantitative real-time PCR in bamboo (Phyllostachys edulis). PLoS ONE 8, e56573. https://doi.org/10.1371/journal.pone.0056573 (2013).
Wang, H. B., Chen, S. M., Jiang, J. F., Zhang, F. & Chen, F. D. Reference gene selection for cross-species and cross-ploidy level comparisons in Chrysanthemum spp. Sci. Rep. 5, 8094. https://doi.org/10.1038/srep08094 (2015).
Garrido, J., Aguilar, M. & Prieto, P. Identification and validation of reference genes for RT-qPCR normalization in wheat meiosis. Sci. Rep. 10, 2726. https://doi.org/10.1038/s41598-020-59580-5 (2020).
Jia, Y. et al. Reference gene selection and validation by qRT-PCR during flower development and in different organs of Primula forbesii. J. Hortic. Sci. Biotech. 95, 383–394. https://doi.org/10.1080/14620316.2019.1681909 (2019).
Tong, Z., Gao, Z., Wang, F., Zhou, J. & Zhang, Z. Selection of reliable reference genes for gene expression studies in peach using real-time PCR. BMC Mol. Biol. 10, 71. https://doi.org/10.1186/1471-2199-10-71 (2009).
Wang, G. L. et al. Selection of reliable reference genes for quantitative RT-PCR in garlic under salt stress. PeerJ 7, e7319. https://doi.org/10.7717/peerj.7319 (2019).
Li, L., Li, N., Fang, H., Qi, X. & Zhou, Y. Selection and validation of reference genes for normalisation of gene expression in Glehnia littoralis. Sci. Rep. 10, 7374. https://doi.org/10.1038/s41598-020-63917-5 (2020).
Gu, C. S. et al. Reference gene selection for quantitative real-time RT-PCR normalization in Iris. lactea var. chinensis roots under cadmium, lead, and salt stress conditions. Sci. World J. 2014, 532713. https://doi.org/10.1155/2014/532713 (2014).
Gu, G. S. et al. De novo characterization of the Iris lactea var. chinensis transcriptome and an analysis of genes under cadmium or lead exposure. Ecotox Environ. Safe 144, 507–513. https://doi.org/10.1016/j.ecoenv.2017.06.071 (2017).
Taylor, S., Wakem, M., Dijkman, G., Alsarraj, M. & Nguyen, M. A practical approach to RT-qPCR-Publishing data that conform to the MIQE guidelines. Methods 50, S1–S5. https://doi.org/10.1016/j.ymeth.2010.01.005 (2010).
Zhang, H. L. et al. Precocious flowering in trees: the Flowering Locus T gene as a research and breeding tool in Populus. J Exp Bot 61, 2549–2560. https://doi.org/10.1093/jxb/erq092 (2010).
Acknowledgements
The work was supported by the National Natural Science Foundation of China (31801901), the Natural Science Foundation of Jiangsu (BK20180314, BK20180317), the Foundation of Key Laboratory of Landsca** (KF201901), Ministry of Agriculture and Rural Affairs, P. R. China, and the Jiangsu Key Laboratory for the Research and Utilization of Plant Resources (JSPKLB201814).
Author information
Authors and Affiliations
Contributions
Y.W., H.Y. conceived and designed the experiments. Y.W., Y.Z., Q.L., T.Z. and L.L. performed the experiments and collected the data. Y.W., H.T. and S.H. prepared reagents/materials/analysis tools. Y.W. and C.G. analyzed the data; Y.W. wrote the paper. Y.W. and H.Y. revised the paper. All authors read and approved the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons licence, and indicate if changes were made. The images or other third party material in this article are included in the article's Creative Commons licence, unless indicated otherwise in a credit line to the material. If material is not included in the article's Creative Commons licence and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Wang, Y., Zhang, Y., Liu, Q. et al. Selection and validation of appropriate reference genes for RT-qPCR analysis of flowering stages and different genotypes of Iris germanica L. Sci Rep 11, 9901 (2021). https://doi.org/10.1038/s41598-021-89100-y
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41598-021-89100-y
- Springer Nature Limited
This article is cited by
-
Evaluation and validation of suitable reference genes for quantitative real-time PCR analysis in lotus (Nelumbo nucifera Gaertn.)
Scientific Reports (2024)
-
Reference genes for qPCR expression in black tiger shrimp, Penaeus monodon
Molecular Biology Reports (2024)
-
Selection and validation of appropriate reference genes for RT–qPCR analysis of Nitraria sibirica under various abiotic stresses
BMC Plant Biology (2022)
-
Identification of appropriate housekee** genes for gene expression studies in human renal cell carcinoma under hypoxic conditions
Molecular Biology Reports (2022)
-
Validation of reference genes for quantitative real-time PCR in chemical exposed and at different age’s brackish water flea Diaphanosoma celebensis
Scientific Reports (2021)