Abstract
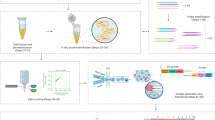
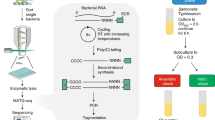
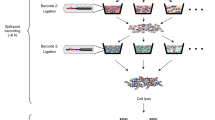
Microbial split-pool ligation transcriptomics (microSPLiT) is a high-throughput single-cell RNA sequencing method for bacteria. With four combinatorial barcoding rounds, microSPLiT can profile transcriptional states in hundreds of thousands of Gram-negative and Gram-positive bacteria in a single experiment without specialized equipment. As bacterial samples are fixed and permeabilized before barcoding, they can be collected and stored ahead of time. During the first barcoding round, the fixed and permeabilized bacteria are distributed into a 96-well plate, where their transcripts are reverse transcribed into cDNA and labeled with the first well-specific barcode inside the cells. The cells are mixed and redistributed two more times into new 96-well plates, where the second and third barcodes are appended to the cDNA via in-cell ligation reactions. Finally, the cells are mixed and divided into aliquot sub-libraries, which can be stored until future use or prepared for sequencing with the addition of a fourth barcode. It takes 4 days to generate sequencing-ready libraries, including 1 day for collection and overnight fixation of samples. The standard plate setup enables single-cell transcriptional profiling of up to 1 million bacterial cells and up to 96 samples in a single barcoding experiment, with the possibility of expansion by adding barcoding rounds. The protocol requires experience in basic molecular biology techniques, handling of bacterial samples and preparation of DNA libraries for next-generation sequencing. It can be performed by experienced undergraduate or graduate students. Data analysis requires access to computing resources, familiarity with Unix command line and basic experience with Python or R.
Key points
-
Through four rounds of combinatorial barcoding, this low-cost and high-throughput single-cell RNA sequencing method enables transcriptomic analysis of individual bacterial cells and the detection of phenotypically distinct subpopulations.
-
Conditions for cell wall digestion, membrane permeabilization and the barcoding procedure are optimized to profile tens of thousands of bacterial cells in a single bench-based barcoding experiment without the need for a dedicated instrument.





Similar content being viewed by others
Data availability
The main data discussed in this protocol are available in the supporting primary research publication (https://doi.org/10.1126/science.aba5257). The raw sequencing files are available at the Sequence Read Archive: GSM4594094, GSM4594095 and GSM4594096. Processed data were submitted to Gene Expression Omnibus, with accession number GSE151940.
References
Raj, A. & van Oudenaarden, A. Nature, nurture, or chance: stochastic gene expression and its consequences. Cell 135, 216–226 (2008).
Avery, S. V. Microbial cell individuality and the underlying sources of heterogeneity. Nat. Rev. Microbiol. 4, 577–587 (2006).
Leisner, M., Stingl, K., Frey, E. & Maier, B. Stochastic switching to competence. Curr. Opin. Microbiol. 11, 553–559 (2008).
Arnoldini, M. et al. Bistable expression of virulence genes in Salmonella leads to the formation of an antibiotic-tolerant subpopulation. PLoS Biol. 12, e1001928 (2014).
Rosenthal, A. Z. et al. Metabolic interactions between dynamic bacterial subpopulations. eLife 7, e33099 (2018).
Real, E. et al. A single-cell atlas of Plasmodium falciparum transmission through the mosquito. Nat. Commun. 12, 1–13 (2021).
Macosko, E. Z. et al. Highly parallel genome-wide expression profiling of individual cells using nanoliter droplets. Cell 161, 1202–1214 (2015).
Klein, A. M. et al. Droplet barcoding for single-cell transcriptomics applied to embryonic stem cells. Cell 161, 1187–1201 (2015).
Jones, R. C. et al. The Tabula Sapiens: a multiple-organ, single-cell transcriptomic atlas of humans. Science 376, eabl4896 (2022).
Blattman, S. B., Jiang, W., Jiang, W., Oikonomou, P. & Tavazoie, S. Prokaryotic single-cell RNA sequencing by in situ combinatorial indexing. Nat. Microbiol. 5, 1192–1201 (2020).
Kuchina, A. et al. Microbial single-cell RNA sequencing by split-pool barcoding. Science 371, eaba5257 (2020).
Wendisch, V. F. et al. Isolation of Escherichia coli mRNA and comparison of expression using mRNA and total RNA on DNA microarrays. Anal. Biochem. 290, 205–213 (2001).
Kaminow, B., Yunusov, D. & Dobin, A. STARsolo: accurate, fast and versatile map**/quantification of single-cell and single-nucleus RNA-seq data. Preprint at https://www.biorxiv.org/content/10.1101/2021.05.05.442755v1 (2021).
Wolf, F. A., Angerer, P. & Theis, F. J. SCANPY: large-scale single-cell gene expression data analysis. Genome Biol. 19, 15 (2018).
Satija, R., Farrell, J. A., Gennert, D., Schier, A. F. & Regev, A. Spatial reconstruction of single-cell gene expression data. Nat. Biotechnol. 33, 495–502 (2015).
Rosenberg, A. B. et al. Single-cell profiling of the develo** mouse brain and spinal cord with split-pool barcoding. Science 360, 176–182 (2018).
Bagnoli, J. W. et al. Sensitive and powerful single-cell RNA sequencing using mcSCRB-seq. Nat. Commun. 9, 2937 (2018).
Nadezhdin, E., Murphy, N., Dalchau, N., Phillips, A. & Locke, J. C. W. Stochastic pulsing of gene expression enables the generation of spatial patterns in Bacillus subtilis biofilms. Nat. Commun. 11, 1–12 (2020).
Dar, D., Dar, N., Cai, L. & Newman, D. K. Spatial transcriptomics of planktonic and sessile bacterial populations at single-cell resolution. Science 373, eabi4882 (2021).
Stoodley, P., Sauer, K., Davies, D. G. & Costerton, J. W. Biofilms as complex differentiated communities. Annu. Rev. Microbiol. 56, 187–209 (2003).
Balaban, N. Q., Merrin, J., Chait, R., Kowalik, L. & Leibler, S. Bacterial persistence as a phenotypic switch. Science 305, 1622–1625 (2004).
Stapels, D. A. C. et al. Salmonella persisters undermine host immune defenses during antibiotic treatment. Science 362, 1156–1160 (2018).
Nguyen, H., Tran, D., Tran, B., Pehlivan, B. & Nguyen, T. A comprehensive survey of regulatory network inference methods using single cell RNA sequencing data. Brief. Bioinform. 22, 1–15 (2021).
Wang, B. et al. Single-cell massively-parallel multiplexed microbial sequencing (M3-seq) identifies rare bacterial populations and profiles phage infection. Nat. Microbiol. 8, 1846–1862 (2023).
Ma, P. et al. Bacterial droplet-based single-cell RNA-seq reveals antibiotic-associated heterogeneous cellular states. Cell 186, 877–891.e14 (2023).
Mcnulty, R. et al. Probe-based bacterial single-cell RNA sequencing predicts toxin regulation. Nat. Microbiol. 8, 934–945 (2023).
Xu, Z. et al. Droplet-based high-throughput single microbe RNA sequencing by smRandom-seq. Nat. Commun. 14, 1–12 (2023).
Imdahl, F., Vafadarnejad, E., Homberger, C., Saliba, A. E. & Vogel, J. Single-cell RNA-sequencing reports growth-condition-specific global transcriptomes of individual bacteria. Nat. Microbiol. 5, 1202–1206 (2020).
Espejo, R. T. & Plaza, N. Multiple ribosomal RNA operons in bacteria; their concerted evolution and potential consequences on the rate of evolution of their 16S rRNA. Front. Microbiol. 9, 338498 (2018).
Dobin, A. et al. STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013).
Homberger, C., Barquist, L. & Vogel, J. Ushering in a new era of single-cell transcriptomics in bacteria. Microlife 3, uqac020 (2022).
Martin, B. K. et al. Optimized single-nucleus transcriptional profiling by combinatorial indexing. Nat. Protoc. 18, 188–207 (2022).
Luecken, M. D. & Theis, F. J. Current best practices in single-cell RNA-seq analysis: a tutorial. Mol. Syst. Biol. 15, e8746 (2019).
Schnabel, Z. E. The estimation of the total fish population of a lake. Am. Math. Mon. 45, 348–352 (1938).
Acknowledgements
We thank Nikolay Burnaevskiy for help with setting up the STARsolo workflow. A.K. and G.S. acknowledge support from the Department of Energy Office of Science, Biological and Environmental Research (BER) Program, Grant DE-SC0023091. A.K. is supported by the National Institute of Dental and Craniofacial Research of the National Institutes of Health (NIH) Grant R21DE032890 and from the National Institute of General Medical Sciences of the NIH Grant R35GM150994.
Author information
Authors and Affiliations
Contributions
K.D.G., S.N.S. and A.K. tested and formalized the microSPLiT protocol and produced the presented figures. L.M.B., L.P., R.W.D., G.S. and A.K. primarily developed the microSPLiT method. K.D.G., S.N.S., C.M.R., A.B.R and M.H. optimized sequencing library preparation. K.D.G. and A.K. implemented the sequencing data analysis workflow. K.D.G., S.N.S. and A.K. wrote the manuscript. A.K. supervised the work.
Corresponding author
Ethics declarations
Competing interests
A.K., L.M.B., R.W.D. and G.S. are inventors on a patent application for microSPLiT filed by the University of Washington. C.M.R., A.B.R and G.S. are cofounders and shareholders of Parse Biosciences, an scRNA-seq company.
Peer review
Peer review information
Nature Protocols thanks Anne-Kristin Kaster and Lars Barquist for their contribution to the peer review of this work.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Related links
Key reference using this protocol
Kuchina, A. et al. Science 371, eaba5257 (2020): https://doi.org/10.1126/science.aba5257
Extended data
Extended Data Fig. 1 Schematic of a microSPLiT sequencing construct.
Only the polyA-primed construct is shown as an example. The random hexamer-primed construct would harbor six random nucleotides in place of the dT15 sequence.
Extended Data Fig. 2 Example data before filtering, shown as a barcode-rank plot.
A red line indicates a chosen threshold for filtering out low-quality cells and debris. a, A high-resolution experiment; data were filtered at 400 total UMIs/cell. Before filtering: 884,736 barcode combinations; after filtering: 60,831 combinations, corresponding to cells. b, A poor-resolution experiment; data were filtered at 350 total UMIs/cell. Before filtering: 884,736 barcode combinations; after filtering: 644 combinations.
Supplementary information
Supplementary Tables 1–4
Supplementary Table 1: cost estimate of microSPLiT. Supplementary Table 2: cost estimate and comparison of high-throughput bacterial scRNA-seq methods. Supplementary Table 3: sequences of oligonucleotides and source barcode plates used in microSPLiT. Supplementary Table 4: barcode sequence files for STARsolo analysis.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gaisser, K.D., Skloss, S.N., Brettner, L.M. et al. High-throughput single-cell transcriptomics of bacteria using combinatorial barcoding. Nat Protoc (2024). https://doi.org/10.1038/s41596-024-01007-w
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41596-024-01007-w
- Springer Nature Limited