Abstract
Somatic cell reprogramming and oncogenic transformation share surprisingly similar features, yet transformed cells are resistant to reprogramming. Epigenetic barriers must block transformed cells from reprogramming, but the nature of those barriers is unclear. In this study, we generated a systematic panel of transformed mouse embryonic fibroblasts (MEFs) using oncogenic transgenes and discovered transformed cell lines compatible with reprogramming when transfected with Oct4/Sox2/Klf4/Myc. By comparing the reprogramming-capable and incapable transformed lines we identified multiple stages of failure in the reprogramming process. Some transformed lines failed at an early stage, whilst other lines seemed to progress through a conventional reprogramming process. Finally, we show that MEK inhibition overcomes one critical reprogramming barrier by indirectly suppressing a hyperacetylated active epigenetic state. This study reveals that diverse epigenetic barriers underly resistance to reprogramming of transformed cells.
Similar content being viewed by others
Introduction
Transformed cells and embryonic stem cells (ESCs) have a remarkable list of similarities [1]. Both cell types have a relaxed chromatin state [2], adopt a glycolysis-biased metabolism despite the availability of oxygen [3, 4], can undergo an epithelial-mesenchymal transition (EMT) [5], and form teratomas [6]. Transformed cells acquire features reminiscent of embryonic development, such as increased cellular plasticity and the upregulated expression of pluripotent genes, including OCT4, NANOG, and SOX2 [7]. Indeed, the expression of pluripotent-specific genes in patient tumor samples correlates with poor clinical outcomes [8, 9]. For example, OCT4 expression is associated with germ cell tumors and cancer stem cells [10, 11], SOX2 expression with glioblastoma [12], and NANOG with colorectal [13] and prostate cancer [14].
Somatic cells can be reprogrammed into induced pluripotent stem cells (iPSCs) by the transfection of a cocktail of transgenes, particularly Oct4 (Pou5f1), Sox2, Klf4, and Myc [15]. Curiously, despite being an artificial process, the reprogramming of somatic cells to iPSCs passes through distinct phases, reminiscent of a developmental program [16,17,18,19]. Tumorigenic transformation also passes through a series of distinct phases in a process that has similarities to somatic cell reprogramming [1, 20]. Some studies suggest direct links between reprogramming and cancer. For example, transient in vivo activation of reprogramming factors leads to tumor formation [21, 22], and cancer-associated mutations in the transcription factor SOX17 can confer reprogramming capability to the normally incapable SOX17 [23].
There have been reports on reprogramming primary human cancer cells to an embryonic state, including cancerous cells from the liver, gastrointestinal tract, and sarcoma cell lines [24,25,26,27]. However, the bona fide reprogramming of these lines is not always clear. Blood cancer cells seem more amenable: human KBM7 cells, T cell acute lymphoblastic leukemia, acute myeloid leukemia [28,29,30,31], lymphoblastoid cells [32, 33], and chronic myeloid leukemia cells [27, 34,35,36] all have reports of successful reprogramming. However, the process of reprogramming is inefficient and often incomplete, and transformed cells are resistant to reprogramming [31, 37]. Additionally, it is unclear how closely the iPSC-like cells resemble iPSCs. For example, the overall gene expression of the reprogrammed cancer cells remains distinct from genuine ESCs/iPSCs [26, 32, 38]. A significant problem is the reprogramming of untransformed bystander cells from primary cancer tissues [31, 39]. Finally, the efficiency of reprogramming transformed cells is much lower than wildtype reprogramming. This is a curious contradiction. Considering the similarities between cancer cells and iPSCs and the pathways used to generate them, it seems that reprogramming should be easier in transformed cells as they have reduced barriers to cell-type conversion. A deeper understanding of the relationship between reprogramming and tumorigenic transformation may shed light on mechanisms of cell type control in cancer development. Additionally, reprogramming transformed cells to a normal iPSC-like state can model cancer development as tumorigenesis can be studied not only in the tissue type where the tumor originated but also in other cell types [29, 31]. These models could be used to explore the earliest stages of cancer development that are usually hidden in humans.
To explore the epigenetic barriers blocking the reprogramming of transformed cells, we generated a panel of ten artificial transformed mouse cell lines. Seven of these lines can acquire OCT4-GFP+ reporter expression and pluripotent characteristics using a conventional OSKM-reprogramming protocol, albeit at very low efficiency. Of the remaining three reprogramming-incapable lines, we show that the defects in reprogramming are line-specific. Reprogramming is a phased process and some lines fail in the early phases, others in the later phases. Compared to wildtype cells, the reprogramming-incapable lines show a heightened ‘hyperactive’ chromatin state and demonstrate global increases in chromatin accessibility and histone acetylation. Some of the transformed lines could be converted to reprogramming-capable by inhibiting MEK signaling which moderates the active chromatin state and leads to decreased histone acetylation.
Results
Transformed mouse cell lines are resistant to reprogramming
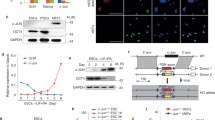
To explore reprogramming in transformed cell lines we first reprogramed established mouse cell lines that were either spontaneously transformed or derived from a primary tumor. We chose three mouse cell lines: 3T3-L1, a spontaneously immortalized fibroblastic cell line; 4T1, a metastatic breast tumor cell line; and N2a (Neuro-2a), a neuroblastoma cell line (Fig. 1a). We attempted to reprogram these cell lines using OSKM in serum+Vc (Vitamin C). Vc was added to all reprogramming conditions, unless otherwise indicated, to accelerate reprogramming [40]. The 3T3-L1 and 4T1 (but not the N2a) cells formed colonies that morphologically resembled iPSCs on day 15 (Fig. 1a). We did not attempt to reprogram for longer than 15 days as the transformed cell lines grow rapidly and outcompeted the morphological iPSC-like colonies. Low levels of NANOG protein could be detected by immunofluorescence in some 3T3-L1 colonies, but not in 4T1 cells (Fig. 1b). We manually picked the iPSC-like colonies to establish iPSC lines. However, contaminating non-reprogrammed 3T3-L1 and 4T1 cells would outcompete iPSC-like colonies, and based on morphology, the cultures would revert to homogenous 3T3-L1 or 4T1 cultures within one passage. To select for reprogramming cells we used a system involving ICAM and CD44 staining as ICAM+/CD44- cells correlate with the expression of NANOG [41]. We detected small numbers of ICAM+/CD44- cells (~1–1.5%) at day 15 of reprogramming in 3T3-L1 and 4T1, but not N2a cells (Fig. 1c, d and Supplementary Table 1 for this and subsequent figures). qRT-PCR of the ICAM+/CD44- cells indicated that Esrrb and Nanog were upregulated in the 4T1, and Essrb, Nanog, and endogenous-Pou5f1 in 3T3-L1 (Fig. 1e). However, when the sorted cells were replated, they rapidly reverted to the original cell type morphology and there was no evidence of iPSC-like cells (Fig. 1f). Whilst there was some evidence of NANOG protein in the 3T3-L1 cells, there was no expression of NANOG in the 4T1, despite some cells being ICAM+/CD44− (Fig. 1b–d). ICAM+/CD44− cells correlate closely with NANOG expression, however, there was not a perfect match in the original study, and some cells remained ICAM+/CD44−/NANOG- [41]. Our reprogrammed cells are likely failing to commit to a pluripotent state. Potentially, reprogramming for longer than 15 days may allow the derivation of iPSC-like cells if appropriate cell culture and sorting conditions could be designed to select reprogrammed cells.
a Bright-field images of the reprogramming of 3T3-L1, 4T1, and N2a cell lines in serum+Vc reprogramming conditions. Images are shown from day 0 (D0) or day 15 (D15) of the reprogramming experiment. Morphologically iPSC-like cells can typically be observed at day 15 and onwards in wildtype reprogramming. Scale bar = 100 µm. b Fluorescent microscopy images for NANOG and tdTomato expressed from the OSKM transgene cassette for the reprogramming of 3T3-L1 and 4T1 cells, from two biological replicates with three images per slide. Scale bar = 50 µm. c Example FACS (fluorescent activated cell sorting) plots for wildtype MEFs and mESCs and cells from a reprogramming time course at day 0 or 15 from 3T3-L1, 4T1, and N2a cells. d Bar chart of the percentage of ICAM+/CD44− sorted cells in the indicated cell lines at day 0 or day 15 of reprogramming. Data is from the mean of three biological replicates. e RT-qPCR showed the indicated gene expression from pooled cultures of the sorted ICAM+ and CD44− cells from the indicated cell lines five days after replating. The mean of four biological replicates, each with three technical replicates ± s.d. is shown. f Bright-field images of the sorted ICAM+ and CD44− cells from the indicated cell lines five days after replating. Scale bar = 100 µm.
Untransformed MEFs senesce and fail to reprogram
Wildtype MEFs can only reprogram at early passages [42], and efficiency declines rapidly before complete failure after passage 4 (Fig. S1a) [43]. RNA-seq of MEFs from passages 1-6 revealed changes in cell cycle genes, including the downregulation of cyclins and other positive cell cycle regulators and the activation of cell cycle inhibitors, particularly Cdkn1a (Fig. S1b). We divided the samples into MEFs that could reprogram successfully (P1, P2) or failed (P5, P6) and measured significantly differentially expressed (DE) genes. In total, 337 genes were significantly upregulated and 419 significantly downregulated (Fig. S1c, d). Gene ontology (GO) and gene set enrichment analysis (GSEA) of the differentially expressed genes indicated that the downregulated genes were related to the cell cycle and extracellular matrix. Upregulated genes were related to cell migration/adhesion, MAPK activity, apoptosis, inflammation, and chemokine expression (Fig. S1e–g). We defined the upregulated gene set as the ‘MEF senescent signature’ and the downregulated gene set as a ‘reprogramming permissive signature’ (Supplementary Table S2).
Generation of a panel of oncogenic transformed MEF cell lines
As the transformed cell lines have been maintained in culture for an extended period, they have accumulated genetic alterations to adapt to the cell culture environment. These changes may permanently impair their ability to reprogram. Consequently, to achieve a controlled system, we generated immortalized MEFs using 10 different combinations of factors to represent a spectrum of transformed cells. The factors were chosen to cover different methods of immortalization: oncogenic transcription factors (Myc, HrasG12V, Mef2d, p53DD), viral transforming factors (SV40T, E1A), anti-apoptotic factors (Bcl2), and an engineered epigenetic factor (Hdac7SA; serines at positions 178, 344, and 479 substituted with alanine, to block nuclear export) [44,45,46] (Supplementary Table S3). Not all of these factors could transform MEFs, and, except for SV40T, at least two factors were required for successful transformation (Fig. S2a, b). The immortalized cell lines were maintained for at least 1 month to remove any non-transformed background MEFs, and the continued expression of transgenes was confirmed by RNA-seq, RT-qPCR, and western blot (Fig. S2c–e). The exception was the p53DD.Myc lines, where Myc could be detected, but p53DD protein could not (Fig. S2c–e). This suggests that both transgenes are required to generate immortal MEFs, but once immortalized, p53DD becomes dispensable and Myc is sufficient.
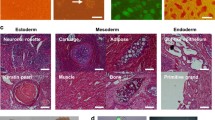
We next looked at the features of the transformed cells. All of the immortalized MEFs proliferated faster than the wildtype MEFs (Fig. S2f). Hras.E1A, SV40T, and Hras.Myc could form tumors when injected into nude (BALB/cNj-Foxn1nu/Gpt) mice, whilst Bcl2.Myc and Hdac7SA.Mef2d could not (Fig. S2g, h). Hematoxylin and eosin staining of cross-sections through the tumors indicated various differentiation layers, although mainly mesoderm (Fig. S2i). Aneuploidy is a common feature of cancer, although its role in transformation remains unclear [47]. A normal karyotype is required for post-implantation embryonic development [48] but may be compatible with pluripotency [49,50,51]. Nonetheless, to rule out the impact of karyotype abnormalities on reprogramming capability we confirmed a normal karyotype for four of the transformed MEF lines that the study will mainly focus on (Fig. S2j).
Transformed cell lines have a spectrum of reprogramming capability
The immortalized MEF cell lines were reprogrammed using a polycistronic lentiviral OSKM system with vitamin C (Vc), to promote reprogramming [43]. We used transformed lines derived from OG2 MEFs, which contain an Oct4-GFP reporter [43]. Surprisingly, several lines generated GFP+ colonies. Based upon the number of GFP+ colonies generated, we labeled the lines as ‘succeeding’ (Hdac7SA.Mef2d, p53DD.Myc), ‘struggling’ (Bcl2.Myc, Hdac7SA.E1A, p53DD.E1A, Hdac7SA.Myc), and ‘failing’ (Bcl2.E1A, Hras.E1A, Hras.Myc, and SV40T) (Fig. 2a, b). Reprogramming the transformed lines was less efficient than wildtype MEFs, except for the p53DD.Myc line. Complete reprogramming requires passages before the pluripotency gene expression program can be stably established. This process partially relies on the fast-dividing iPSCs outcompeting the slow-growing/senescent wild-type MEFs, however, the transformed MEFs were also highly proliferative (Fig. S2f) and would compete with the reprogrammed iPSCs. Hence, we FACS sorted the GFP+ cells and passaged them to establish iPSC lines in the absence of transformed MEFs. In a panel of marker genes (Fig. 2c), the expression of fibroblastic genes was surprisingly noisy in the transformed MEFs, suggesting transformation has a strong effect. The transformed MEFs retained high expression of mesenchymal genes (Cdh2, Snai2, Zeb1, and Zeb2) and low epithelial genes (Epcam, Pecam1, and Cdh1), whilst the reprogrammed iPSC-like cells had the opposite pattern (Fig. 2c). The expression of the pluripotency markers was validated in the iPSC-like lines by immunofluorescence staining (NANOG), RT-qPCR (Esrrb, endogenous-Pou5f1, Nanog), and Western blot (SOX2 and NANOG) (Fig. S3a–c).
a Representative brightfield and GFP images from reprogrammed transformed MEFs for the indicated cell lines. Scale bar = 100 µm. b Chart showing the number of Oct4-GFP+ colonies from the indicated transformed MEF lines. Each MEF line was at least 1 month old. Reprogramming was performed between three and five independent biological replicates (See Supplementary Table 1). Circles represent biological replicates; the red line indicates the mean and the error bars indicate the standard error of the mean (SEM). c Heatmap from the RNA-seq data showing the expression of a representative set of genes specific to somatic fibroblastic cells, pluripotent stem cells, or from the MET (mesenchymal-epithelial transition). NTC=normalized tag count. d Correlation heatmap (R2) of the original MEF samples and the reprogrammed GFP+ cells and ESCs. e PCA scatter plot showing the first two dimensions of the RNA-seq data from the original wildtype or transformed MEF lines (red) and the GFP+ cell lines (blue) and ESCs (green) maintained in serum+LIF or 2i+LIF.
The reprogrammed iPSC-like lines had high levels of pluripotency genes and low levels of somatic genes (Fig. 2c), and cross-correlation and principal component analysis (PCA) indicated the cells were similar to ESCs (Fig. 2d, e). We used DPre, a computational tool that identifies the cell type based on RNA-seq expression [52, 53]. DPre identified most GFP+ lines as ESC-like (Fig. S3d, e). The exception was Bcl2.Myc GFP+ Line #2, which had only a weak ESC-like character (Fig. S3d). Additionally, DPre also indicated that several GFP+ lines (Hdac7SA.Mef2d GFP+ line #4, Bcl2.E1A Vc GFP+ line #1, Bcl2.Myc GFP+ line #3) were contaminated with transformed MEFs (Fig. S3d, e). Normal reprogramming relies upon untransformed MEFs senescing and being outcompeted by rapidly growing iPSCs. However, the transformed MEFs can grow as fast as iPSCs and no longer senesce, meaning contamination of iPSC cultures remains a problem.
We confirmed that the iPSC lines could form teratomas with tissues representing all three germ layers (Fig. S3f). A mark of complete reprogramming is the silencing of the OSKM transgenes, and the OSKM transgene was not detected in RNA-seq data (Fig. S3g). Interestingly, the immortalization factors were only partially silenced. All were silenced except E1A in the Bcl2.E1A GFP+ line and Myc in the Bcl2.Myc GFP+ lines (Fig. S3g). We confirmed Hdac7SA and Mef2d were silenced by qRT-PCR (Fig. S3h). Overall, these data confirmed the successful generation of iPSC-like lines from some transformed tumorigenic MEFs.
Transformed MEFs have multiple transcriptional perturbations
We next explored the properties that made some immortal MEFs reprogramming-capable and others –incapable. We focused on two (non-exclusive) models: the inability to reprogram occurs due to problems in the originating MEFs, or it is caused by failures to traverse the correct sequence of events required for successful reprogramming.
Immortalization of somatic adult cells occurs by a range of mechanisms and is accompanied by changes in the epigenetic state and gene expression patterns. The RNA-seq data of the transformed MEFs showed substantial changes in gene expression and a large variation versus wildtype MEFs (Figs. 2c, e, and 3a). There was no simple correlation between the number of gene changes and reprogramming capability (Fig. 3a). Despite a large number of gene expression changes, there was no evidence that the MEFs were transdifferentiating as their global gene expression patterns continued to correlate well against wildtype MEFs (Fig. 3b), and DPre continued to identify the cells as MEF-like (Fig. S4a). Gene ontology of the DE genes suggested changes were associated with basic cellular processes such as upregulation of metabolic processes, extracellular matrix genes, and downregulation of ribosomes, cell cycle, and DNA repair processes (Fig. 3c, S4b). The MEF-senescent gene signature (defined in Fig. S1c) was not upregulated in the transformed lines, and the majority of lines had a significantly reduced MEF-senescent signature, reminiscent of the level in ESCs (Fig. 3d). This indicates the transformed lines are avoiding senescence. Interestingly, the reprogramming-competent signature (defined in Fig. S1c) declines in all lines (except SV40T), which suggests a reduced capability to reprogram (Fig. 3d).
a Chart showing the number of significantly differentially regulated genes up (+) or down (−) regulated (Bonferroni-Hochberg corrected p-value of <0.01 and a minimum fold-change of 2). b Correlation (R2) of the immortalized MEF lines compared to the wildtype MEFs. c Selected GO terms from the significantly up or downregulated genes from the transformed MEF lines. A term was considered significant if it had a Bonferroni-Hochberg corrected p-value of <0.01). See also Fig. S4b for the full table. d Boxplots for the genes defined as the MEF-senescent signature or the reprogramming-capable signature from Fig. S1c. Significance is from a two-sided Welch’s t-test for pair-wise comparisons versus MEFICR wildtype Passage 2 as the reference. A change was considered significant if the p-value was <0.01. Significant changes are colored red or blue.
Surprisingly, the patterns of the gene expression changes were uniform across cell lines; genes up or downregulated in one transformed line tended to be either unchanged or similarly deregulated in other lines (Fig. S4c). This implies a shared transformation signature.
We next looked at specific genes identified as key regulators of the reprogramming process. Three factors related to cancer and cellular transformation that modulate reprogramming in wildtype cells are Tp53, Cdkn1a, and Rb1 (Retinoblastoma) [45, 54,55,56,57]. There was little change in Rb1 mRNA levels, and whilst Cdkn1a was relatively high in all cell lines except p53DD.E1A, SV40T, and wildtype MEFs, its expression was not correlated with reprogramming capability (Fig. S5a). For Tp53, its expression was relatively consistent across the MEF lines and did not correlate with reprogramming ability (Fig. S5a). We inferred p53 activity by looking at known target genes of p53, and all lines showed unaltered p53 activity except for the MEF line containing the dominant-negative p53DD (p53DD.E1A). This line had significantly downregulated p53 target genes (Fig. S5b–c), reduced p53 phosphorylation, and reduced MDM2 protein levels (Fig. S5d). Loss of p53 is beneficial for reprogramming in wildtype cells [45, 57, 58], yet in transformed lines reduced p53 activity in the p53DD.E1A line did not correlate with efficient reprogramming (Fig. 2b). This suggests that, in contrast to wildtype cells, reduced p53 activity is not beneficial for reprogramming in transformed cells.
Transformed MEFs encounter multiple roadblocks at different phases of reprogramming
Reprogramming occurs in defined phases [16, 17, 59], so we explored if there are also changes in the phases of reprogramming in the transformed lines. We performed RNA-seq during reprogramming in the early phase (D3, D6), the mid-phase (D9, D12), and the late phase (D15). These time points roughly correspond to three waves of gene expression labeled initiation, maturation, and stabilization [17, 59]. The same waves were observed in our data, based on sets of genes specific to the three phases (Fig. 4a, S6a). Interestingly, different transformed lines completed different phases of reprogramming and appeared to fail at distinct stages. Lines that can successfully be reprogrammed (Hdac7SA.Mef2d, Bcl2.Myc, and Hdac7SA.Myc) closely resembled the wildtype MEF reprogramming process (Fig. 4a). Hdacs7SA.E1A and p53DD.E1A progressed through maturation but struggled to upregulate stabilization phase genes (Fig. 4a), although both can ultimately generate small numbers of GFP+ cells (Fig. 2b). The remaining transformed lines failed to reprogram at different stages. SV40T and Hras.E1A failed to upregulate maturation genes, but completed initiation, whilst Hras.Myc failed maturation and stabilization (Fig. 4a, S6).
a Line charts showing the expression of initiation, maturation, and stabilization genes [59], at the indicated time points or in MEFs and GFP+ cells. See Fig. S6 for the expression of the individual genes in each class. The red line is the mean of all genes, the grey lines are the individual genes. b Heatmap of the expression of mesenchymal and epithelial marker genes in starting lines and ESCs. c Bar charts showing the number of significantly differentially regulated genes at each time point for the indicated transformed lines versus a wildtype reprogramming time course. Differential expression was compared for each line and time point versus the corresponding wildtype timepoint. A gene was considered differentially regulated if the q-value (Bonferroni-Hochberg corrected p-value) was <0.01 and the fold-change >2. d Heatmaps of significantly overrepresented gene ontology (biological process) terms for downregulated genes from the indicated time points and cell lines.
The mesenchymal-epithelial transition (MET) is a key part of the early stage of reprogramming [59, 60]. All of the cell lines expressed mesenchymal marker genes (Fig. 4b), and all transformed lines managed to navigate the MET, except for Hras.Myc, and to a lesser extent SV40T (Fig. S6b). Interestingly, although Hras.E1A completed the MET, it started with premature upregulation of epithelial genes, for example, Epcam (Fig. 4b, S6b). Considering that the three lines that fail to reprogram, Hras.E1A, Hras.Myc and SV40T all have MET progression problems suggests that disrupting the MET impairs the reprogramming of transformed cells.
We next looked at changes in the overall phases of reprogramming in the early, middle, and late stages of reprogramming. We measured the number of significantly deregulated genes at each time point by performing differential expression versus the WT reprogramming on the same day. This can act as a proxy score for the divergence from a typical reprogramming time course. In this analysis, downregulated genes represent genes that fail to upregulate at the correct time point, whilst upregulated genes represent those genes that are erroneously high during reprogramming (Fig. 4c). As expected, the lines that reprogrammed at the highest efficiency (p53DD.Myc) also had the lowest overall divergence, and only diverged from WT reprogramming at day 0 (Fig. 4c). Lines that could reprogram, but at low efficiency (Hdac7SA.Mef2d, Bcl2.Myc, Hdac7SA.E1A, and Hdac7SA.Myc) tended to have high initial divergence from the MEF state (day 0 and 3), but would correctly regulate reprogramming-associated genes at the later time points (day 12 and 15). Finally, lines that fail to reprogram (Hras.E1A and Hras.Myc), or reprogram exceptionally rarely (Bcl2.E1A), tended to have high divergence at all time points (days 0-15). The exception to these patterns was the SV40T line, which followed the reprogramming gene expression program closely, but would diverge at day 12 (Fig. 4c). Interestingly, GO analysis of the downregulated genes (genes that should be upregulated at that specific time point) highlighted the regulation of DNA repair genes, which were defective in both Hras.E1A and SV40T cell lines (Fig. 4d). This data indicates that transformation-specific effects drive line-specific blocks on reprogramming in a stage-dependent manner.
Chemical intervention can convert some lines from reprogramming-incapable to reprogramming-capable
The analysis above suggested several pathways that could be manipulated to convert reprogramming-incapable to -capable, specifically, the MET, ribosome biogenesis, cell proliferation, cell adhesion, DNA repair, and apoptosis. To attempt to convert reprograming-incapable to -capable, we screened the effect of a range of inhibitors on three reprogramming-capable lines (WT, Hdac7SA.Mef2d, and Bcl2.E1A MEFs) versus three reprogramming-incapable lines (Hras.E1A, Hras.Myc and SV40T MEFs). We chose to exclude p53DD.Myc from this analysis as although it reprogrammed with high efficiency (Fig. 2b), the p53DD transgene was silenced (Fig. S2e), suggesting that its transformation mechanism is complex. In total, we used 25 inhibitors targeting a range of pathways (Fig. S7a). Whilst most inhibitors had no effect or would ablate reprogramming, 5 inhibitors promoted the reprogramming of one or more of the transformed reprogramming-incapable lines (Fig. S7a). The five small molecules identified targetted: MEK1/2 (PD; PD0325901), GSK3 (CHIR; CHIR99021), ROCK (Y; Y23637), G9a (BIX; BIX-01294), and histone deacetylases (TSA). We tested the inhibitors in combinations and found that the most efficient combination was CHIR, PD, and Y (Fig. 5a, S7b). Interestingly, cocktails including PD and CHIR could also improve wildtype MEF reprogramming, in agreement with previous observations [61]. Most transformed cell lines responded to both PD and CHIR, however, PD-alone was able to convert SV40T and Hras.E1A to reprogramming-capable, as serum+Vc+PD alone resulted in a small number of GFP+ cells (Fig. S7b). Conversely, CHIR, Y, and BIX alone were incapable of converting Hras.E1A MEFs to reprogramming-capable (Fig. S7b). Interestingly the inhibitor cocktails had effects on reprogramming in a line-specific manner, as the addition of PD to the reprogramming cocktail converted the normally reprogramming-capable Bcl2.E1A line to incapable. This highlights the line-specific nature of the reprogramming barriers and shows that overcoming a barrier in one line can potentially initiate a barrier in another transformed line.
a Heatmap showing the number of GFP colonies produced depending upon the reprogramming conditions used, for the indicated transformed lines. Reprogramming experiments were performed between two and five biological replicates for each combination of small molecule inhibitors (See Supplementary Table 1 and Fig. S7b). b Chart showing the number of GFP+ colonies in the indicated immortal cell lines with the inhibitor cocktail. Data are from four biological replicates (MEFOGF WT) or three biological replicates (all other transformed cell lines). c Percentage of cells sorted by FACS at day 12 of the reprogramming assay in the indicated transformed cell lines or wildtype MEFs. Data are from four biological replicates. d Fluorescence microscopy images for NANOG at day 15 of a reprogramming time course for the reprogramming of immortalized ICR-background MEFs cells when treated with the inhibitor cocktail combination Vitamin C (Vc) CHIR99021 (CHIR) PD0325901 (PD) Y23637 (Y) and BIX-01294 (BIX). Scale bar = 50 µm. Images were taken from two biological replicates with three images per slide. e PCA of the RNA-seq from the immortalized MEFs, and the reprogrammed iPSCs and ESCs. The various chemical cocktails used during the reprogramming are abbreviated for clarity. Key: Vc = Vitamin C; P = PD0325901; C = CHIR99021; R = Y23637; B = BIX-01294.
We confirmed the resulting iPSC-like cells derived from Hras.E1A or SV40T MEFs reprogrammed with the PD, CHIR, Y, and BIX adopted a normal morphology, expressed pluripotent marker genes by immunofluorescence, qRT-PCR and Western blot (Fig. 5d, and Fig. S7c, d). We sorted the GFP+ cells and allowed the cells to mature through two passages to establish iPSC lines. Gene expression was measured using RNA-seq, and their gene expression was closely correlated with ESCs by both co-correlation and PCA (Fig. 5e and Fig. S8a, b). The results indicate that transformation-specific pathways are impairing the ability to reprogram. Overall, MEK inhibition by PD was the dominant factor in converting reprogramming-incapable to capable.
Epigenetic defects underly the inability to reprogram
We next focused on three specific cell lines: Hdac7SA.Mef2d, Hras.Myc and Hras.E1A. Hdac7SA.Mef2d could reprogram in serum under normal conditions, whilst Hras.E1A lines were initially resistant to reprogramming (Fig. 2b), but the addition of PD converts it to reprogramming-capable (Fig. 5a–d). Hras.Myc, conversely, could not be converted to reprogramming-capable with any of the conditions we tried (Fig. 5b).
We explored the epigenetic regulation of the transformed MEFs. As a proxy for overall epigenetic activity, we looked at the expression levels of epigenetic factors involved in activation, repression, and the reading of epigenetic marks, as defined by the Epifactors database [62]. As expected, epigenetic-related factors were uniformly significantly upregulated in ESCs compared to MEFs (Fig. S9a), reflecting their more complex epigenetic regulation [63]. For the transformed MEFs, activators were more often significantly upregulated, compared to untransformed MEFs (Fig. S9a). Erasers and readers were only upregulated in several transformed lines, all containing E1A: Hras.E1A and Bcl2.E1A and p53DD.E1A (Fig. S9a). Nonetheless, whilst the different classes of epigenetic regulators varied across the transformed lines and did not discriminate reprogramming-capable from incapable, the general pattern for the majority of lines was increased expression of epigenetic regulators.
Western blot of repressive histone modifications (e.g. H3K27me3, H3K9me3) tended to stay the same in the different lines (Fig. 6a). However, histone acetylation inversely correlated with reprogramming capability as H3K27ac, H3ac, or H4ac was high in reprogramming-incapable lines (Hras.Myc, Hras.E1A and SV40T). This result is somewhat contradictory. Acetylation of histones leads to relaxed chromatin and higher gene expression and is overall beneficial for reprogramming [46, Teratoma formation 5–10 million mouse iPSCs in a slurry of Matrigel and mTeSR (1:1) medium were injected into 8-week-old female nude (BALB/cNj-Foxn1nu/Gpt) immunodeficient mice. Teratoma growth was quantified by measuring the approximate elliptical area (mm2) with calipers measuring the outward width and height after growth for 60 days. Representative tumors were dissected and sectioned and slices were stained with hematoxylin and eosin. Reprogrammed cells were stained with alkaline phosphatase according to the manufacturer’s protocols. In brief, reprogramming cells were fixed in 1% (w/v) formaldehyde, and then cells were stained with BCIP/NBT Alkaline Phosphatase Color Development Kit (C3206, Beyotime Biotech) according to the kit’s instructions. Reprogramming cells were digested with trypsin and washed with DPBS once, and analyzed or sorted with a BD FACS Aria III flow cytometer. To monitor cellular reprogramming status, reprogrammed cells were stained with antibodies: anti-CD44-PE (25-0441-82, 1:500, Thermo Fisher) and anti-ICAM-1-FITC (sc-8439 FITC, 1:200, Santa Cruz Biotechnology). Cells were sorted and analyzed on a BD FACS Aria III instrument, with the following gain settings: For cancer cell line (Fig. 1c): FSC 128; SSC 254; FITC 495; PE-cy7 478. For immortalized MEFs GFP detection (Fig. 5c) FSC 166; SSC 263; FITC 310. Cells were lysed with RIPA buffer (P0013B, Beyotime Biotech). Proteins were resolved in sodium dodecyl sulfate-polyacrylamide gel electrophoresis (SDS-PAGE; 15% (w/v) for blots involving histones and 12% (w/v) for all other proteins) and transferred onto pre-activated polyvinylidene fluoride (PVDF) membranes (IPVH00010, Millipore, MA, USA). The PVDF Membranes were incubated with anti-H3 (ab1791, 1:3000, Abcam), anti-H3K27ac (ab4279, 1:2000, Abcam), anti-H3ac (06-599,1:2000, Millipore), anti-H4ac (ab46983, 1:2000, Abcam), anti-H3K4me3 (ab8580, 1:2000, Abcam), anti-H3K4me (ab176877,1:2000, Abcam), anti-H3K27me3 (07-449,1:2000, Millipore), anti-H3K9me3 (ab8898, 1:2000, Abcam), anti-NANOG (8822 S, 1:1000, cell signaling), anti-OCT4 (sc-5279, 1:1000, Santa Cruz Biotechnology), anti-P53 (sc-126, 1:1000, Santa Cruz Biotechnology), anti-HDAC7 (ab12174,1:2000, Abcam), anti-MEF2D (ab32845,1:2000, Abcam), anti-ACTIN (ab8227, 1:3000, Abcam), anti-MYC (ab32074,1:2000, Abcam), anti-SOX2 (4900S, 1:1000, cell signaling), anti-MDM2 (AF7499,1:1000, Beyotime), anti-CNK1A (AF5252,1:1000, Beyotime) anti-Phospho-p53(Ser15) (AF5893,1:1000, Beyotime). Afterward, the membranes were incubated with HRP-conjugated goat antirabbit IgG (ab205718, 1:3000, Abcam) and visualized using an enhanced chemiluminescence BeyoECL method (P0018AS, Beyotime Biotech) on a Tanon 6100C machine. All Uncropped unprocessed western blots are in Supplementary Material. Cells were washed with cold PBS and fixed using 4% (w/v) paraformaldehyde in PBS for 10 min at room temperature. Then cells were washed with PBS and permeabilized with 0.25% (v/v) Triton X-100 for 10 minutes at room temperature. Fixed cells were then washed with PBS three times and blocked with 3% (w/v) bovine serum albumin for 1 hour at room temperature. Then cells were incubated with primary antibodies: anti-NANOG (8822 S, 1:1000, cell signaling), at 4 °C overnight in primary antibody. Following overnight incubation, cells were washed with wash buffer (0.1% (v/v) Tween-20 in PBS) and incubated with secondary antibodies Alexa Fluor series (Life Technologies) in wash buffer for 1 h at room temperature. Cells were washed again and nuclei were stained using DAPI (5 μg/ml, Thermo). Then we used a fluorescent microscope (LSM980 Carl Zeiss) to capture at least three slides for each sample at ×100 magnification. Laser lines 488, 568, and 405 nm were used to stimulate and observe Nanog, tdTomato, and DAPI. Immunofluorescence imaging experiments were performed in biological duplicate, and at least three views were gathered per slide Total RNA from cells was isolated using RNAzol RT (MRC, RN190) according to the manufacturer’s protocols. cDNA synthesis by using a PrimeScript RT Master Mix (Takara, RR036A). Real-time PCR was performed in triplicate using SYBR Premix Ex Taq (Takara, RR820A) and using a Biorad Real-time PCR system. The primers used are listed in Supplementary Table S3. RNA was isolated using RNAzol RT (MRC, RN190) according to the manufacturer’s protocol and prepared for sequencing with RNA-seq NEB Next Ultra RNA Library Prep Kit (NEB, #7530). RNA quality was tested using an Agilent 2100 and a minimum RIN score of 8.0 was required for sequencing. Samples were sequenced on an Illumina Novaseq 6000. RNA-seq was analyzed essentially as described in [52], except scTE/te_counts was used to assign mapped reads to genes and TEs [76], and the UCSC genome browser repeat mask track and GENCODE vM23 annotation was used for TE and gene assignment. Differential expression was determined using DESeq2 [77]. A gene was considered differentially expressed had an absolute fold-change of at least 2, and a Bonferroni-Hochberg corrected p-value of 0.01. GSEA was performed using fgsea [78], and GO was done with goseq [79]. Cell type determination was scored with DPre [53]. Other analyses were performed using glbase3 [80]. ATAC-seq library was generated using the Tn5 enzyme from the TruePrep DNA Library Prep Kit V2 (Vazyme, TD501-02) as previously described [81]. Briefly, a total of ~50,000 cells were washed once with 50 μl of cold PBS and resuspended in 50 μl lysis buffer (10 mM Tris-HCl pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.2% (v/v) IGEPAL CA-630). The nuclei were centrifuged for 10 min at 500 × g at 4 °C, followed by the addition of 50 μl transposition reaction mix (25 μl TD buffer, 2.5 μl Tn5 transposase (Vazyme), and 22.5 μl nuclease-free water). Samples were PCR-amplified and purified using a MinElute kit (Qiagen). After selecting an appropriate PCR cycle number (See [81]) samples were sequenced on an Illumina sequencer. ATAC-seq data was analyzed essentially as described in reference: [66]. Briefly, reads were aligned to the mm10 mouse genome with bowtie2 [82], peaks were called with MACS2 [83], and then the redefine_peaks function, which is a generalized reimplementation of the algorithm in [66], was used to recover low-scoring peaks by sharing peak information across samples [84]. DNA binding motifs were detected using HOMER [85]. All other analyses were performed using glbase3 [80]. ATAC-seq data from GSE93029 [66] and GSE103980 [86] were reanalyzed as part of this study. No statistical test was used to determine the sample size. Animal experiments were performed in biological duplicates, and there was no randomization performed to select animals for experimentation. The investigator was not blinded to the experimental details. Differential gene expression was calculated using DESeq2 (v1.36.0). A gene was considered significantly differentially regulated if it had an absolute fold-change of at least 2 and a Bonferroni-Hochberg corrected p-value (q-value) of <0.01. This criterion was used in Figs. 3a, 4c, Figs. S1c, and d. RNA-seq experiments were performed in at least biological duplicate (different samples on different days, or independent cell lines). Gene ontology analysis was performed using goseq (v1.48.0) and statistics were calculated using goseq’s internal statistical model. A gene ontology category was considered significantly enriched if there were at least 50 genes in that GO term and a Bonferroni-Hochberg corrected p-value (q-value) of <0.01. GSEA was performed using fgsea (v1.22.0). Gene sets were considered enriched or depleted if they had an absolute NES (normalized enrichment score) of at least 1.5 and a Bonferroni-Hochberg corrected p-value (q-value) of <0.01. Transcription factor motif analysis was performed using HOMER. A motif was considered significantly enriched if the uncorrected p-value was <0.00001. Western blots were repeated at least twice with similar results, except for Fig. S7c which was performed once. Figure 6a was repeated three times. FACs analysis (Figs. 1c and 5c) were performed three times with similar results. All qRT-PCR experiments were performed using at least three biological replicates with three technical replicates each. Immunofluorescence imaging was performed in biological duplicate, and at least three views were gathered per slide. In total we generated twenty distinct transformed MEF lines, using ten transgene combinations, in two genetic backgrounds, MEFOG2 and MEFICR. We reprogrammed the MEF OG2 transformed cells to generate 37 iPSC-like lines.Alkaline phosphatase staining
Flow cytometry
Western blot
Immunofluorescence
RT-qPCR
RNA-seq preparation and analysis
ATAC-seq preparation and analysis
Statistical analysis, reporting, and biological replication
Data availability
The RNA-seq and ATAC-seq data generated as part of this study are available in the gene expression omnibus (GEO) public database under accession number GSE213225.
References
Iglesias JM, Gumuzio J, Martin AG. Linking pluripotency reprogramming and cancer. Stem Cells Transl Med. 2017;6:335–9.
Sun L, Fu X, Ma G, Hutchins AP. Chromatin and epigenetic rearrangements in embryonic stem cell fate transitions. Front Cell Dev Biol. 2021;9:637309.
Cha Y, Han MJ, Cha HJ, Zoldan J, Burkart A, Jung JH, et al. Metabolic control of primed human pluripotent stem cell fate and function by the miR-200c-SIRT2 axis. Nat Cell Biol. 2017;19:445–56.
Kubicka A, Matczak K, Labieniec-Watala M. More than meets the eye regarding cancer metabolism. Int J Mol Sci. 2021;22:9507.
Li Q, Hutchins AP, Chen Y, Li S, Shan Y, Liao B, et al. A sequential EMT-MET mechanism drives the differentiation of human embryonic stem cells towards hepatocytes. Nat Commun. 2017;8:15166.
Cunningham JJ, Ulbright TM, Pera MF, Looijenga LH. Lessons from human teratomas to guide development of safe stem cell therapies. Nat Biotechnol. 2012;30:849–57.
Niu N, Mercado-Uribe I, Liu J. Dedifferentiation into blastomere-like cancer stem cells via formation of polyploid giant cancer cells. Oncogene. 2017;36:4887–900.
Hepburn AC, Steele RE, Veeratterapillay R, Wilson L, Kounatidou EE, Barnard A, et al. The induction of core pluripotency master regulators in cancers defines poor clinical outcomes and treatment resistance. Oncogene. 2019;38:4412–24.
Jang HS, Shah NM, Du AY, Dailey ZZ, Pehrsson EC, Godoy PM, et al. Transposable elements drive widespread expression of oncogenes in human cancers. Nat Genet. 2019;51:611–7.
Koo BS, Lee SH, Kim JM, Huang S, Kim SH, Rho YS, et al. Oct4 is a critical regulator of stemness in head and neck squamous carcinoma cells. Oncogene. 2015;34:2317–24.
Rijlaarsdam MA, van Herk HA, Gillis AJ, Stoop H, Jenster G, Martens J, et al. Specific detection of OCT3/4 isoform A/B/B1 expression in solid (germ cell) tumours and cell lines: confirmation of OCT3/4 specificity for germ cell tumours. Br J Cancer. 2011;105:854–63.
Lopez-Bertoni H, Johnson A, Rui Y, Lal B, Sall S, Malloy M, et al. Sox2 induces glioblastoma cell stemness and tumor propagation by repressing TET2 and deregulating 5hmC and 5mC DNA modifications. Signal Transduct Target Ther. 2022;7:37.
Zhang J, Espinoza LA, Kinders RJ, Lawrence SM, Pfister TD, Zhou M, et al. NANOG modulates stemness in human colorectal cancer. Oncogene. 2013;32:4397–405.
Jeter CR, Liu B, Liu X, Chen X, Liu C, Calhoun-Davis T, et al. NANOG promotes cancer stem cell characteristics and prostate cancer resistance to androgen deprivation. Oncogene. 2011;30:3833–45.
Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–76.
Xu Y, Zhang M, Li W, Zhu X, Bao X, Qin B, et al. Transcriptional control of somatic cell reprogramming. Trends Cell Biol. 2016;26:272–88.
Polo JM, Anderssen E, Walsh RM, Schwarz BA, Nefzger CM, Lim SM, et al. A molecular roadmap of reprogramming somatic cells into iPS cells. Cell. 2012;151:1617–32.
Hussein SM, Puri MC, Tonge PD, Benevento M, Corso AJ, Clancy JL, et al. Genome-wide characterization of the routes to pluripotency. Nature. 2014;516:198–206.
Guo L, Lin L, Wang X, Gao M, Cao S, Mai Y, et al. Resolving cell fate decisions during somatic cell reprogramming by single-cell RNA-Seq. Mol Cell. 2019;73:815–29.e7.
Huyghe A, Furlan G, Schroeder J, Cascales E, Trajkova A, Ruel M, et al. Comparative roadmaps of reprogramming and oncogenic transformation identify Bcl11b and Atoh8 as broad regulators of cellular plasticity. Nat Cell Biol. 2022;24:1350–1363.
Abad M, Mosteiro L, Pantoja C, Canamero M, Rayon T, Ors I, et al. Reprogramming in vivo produces teratomas and iPS cells with totipotency features. Nature. 2013;502:340–5.
Ohnishi K, Semi K, Yamamoto T, Shimizu M, Tanaka A, Mitsunaga K, et al. Premature termination of reprogramming in vivo leads to cancer development through altered epigenetic regulation. Cell. 2014;156:663–77.
Srivastava Y, Tan DS, Malik V, Weng M, Javed A, Cojocaru V, et al. Cancer-associated missense mutations enhance the pluripotency reprogramming activity of OCT4 and SOX17. FEBS J. 2020;287:122–44.
Miyoshi N, Ishii H, Nagai K, Hoshino H, Mimori K, Tanaka F, et al. Defined factors induce reprogramming of gastrointestinal cancer cells. Proc Natl Acad Sci USA. 2010;107:40–5.
Kim HJ, Jeong J, Park S, ** YW, Lee SS, Lee SB, et al. Establishment of hepatocellular cancer induced pluripotent stem cells using a reprogramming technique. Gut Liver. 2017;11:261–9.
Zhang X, Cruz FD, Terry M, Remotti F, Matushansky I. Terminal differentiation and loss of tumorigenicity of human cancers via pluripotency-based reprogramming. Oncogene. 2013;32:2249–60. 60 e1-21.
Carette JE, Pruszak J, Varadarajan M, Blomen VA, Gokhale S, Camargo FD, et al. Generation of iPSCs from cultured human malignant cells. Blood. 2010;115:4039–42.
Li T, Zhang Y, Li Y, Wang X, Bao W, Huang J, et al. Modeling leukemia with pediatric acute leukemia patient-derived iPSCs. Stem Cell Res. 2021;54:102404.
Dannenmann B, Klimiankou M, Oswald B, Solovyeva A, Mardan J, Nasri M, et al. iPSC modeling of stage-specific leukemogenesis reveals BAALC as a key oncogene in severe congenital neutropenia. Cell Stem Cell. 2021;28:906–22.e6.
Liu Y, Cheng H, Gao S, Lu X, He F, Hu L, et al. Reprogramming of MLL-AF9 leukemia cells into pluripotent stem cells. Leukemia. 2014;28:1071–80.
Golubeva D, Porras DP, Doyle M, Reid JC, Tanasijevic B, Boyd AL, et al. Reprogramming of acute myeloid leukemia patients cells: harboring cancer mutations requires targeting of AML hierarchy. Stem Cells Transl Med. 2023;12:334–354.
Kumar S, Curran JE, Glahn DC, Blangero J. Utility of lymphoblastoid cell lines for induced pluripotent stem cell generation. Stem Cells Int. 2016;2016:2349261.
Barrett R, Ornelas L, Yeager N, Mandefro B, Sahabian A, Lenaeus L, et al. Reliable generation of induced pluripotent stem cells from human lymphoblastoid cell lines. Stem Cells Transl Med. 2014;3:1429–34.
Bedel A, Pasquet JM, Lippert E, Taillepierre M, Lagarde V, Dabernat S, et al. Variable behavior of iPSCs derived from CML patients for response to TKI and hematopoietic differentiation. PLoS ONE. 2013;8:e71596.
Hu K, Yu J, Suknuntha K, Tian S, Montgomery K, Choi KD, et al. Efficient generation of transgene-free induced pluripotent stem cells from normal and neoplastic bone marrow and cord blood mononuclear cells. Blood. 2011;117:e109–19.
Kumano K, Arai S, Hosoi M, Taoka K, Takayama N, Otsu M, et al. Generation of induced pluripotent stem cells from primary chronic myelogenous leukemia patient samples. Blood. 2012;119:6234–42.
Bang JS, Choi NY, Lee M, Ko K, Park YS, Ko K. Reprogramming of cancer cells into induced pluripotent stem cells questioned. Int J Stem Cells. 2019;12:430–9.
Stricker SH, Feber A, Engstrom PG, Caren H, Kurian KM, Takashima Y, et al. Widespread resetting of DNA methylation in glioblastoma-initiating cells suppresses malignant cellular behavior in a lineage-dependent manner. Genes Dev. 2013;27:654–69.
Lee JH, Salci KR, Reid JC, Orlando L, Tanasijevic B, Shapovalova Z, et al. Brief Report: human acute myeloid leukemia reprogramming to pluripotency is a rare event and selects for patient hematopoietic cells devoid of leukemic mutations. Stem Cells. 2017;35:2095–102.
Esteban MA, Wang T, Qin B, Yang J, Qin D, Cai J, et al. Vitamin C enhances the generation of mouse and human induced pluripotent stem cells. Cell Stem Cell. 2010;6:71–9.
O'Malley J, Skylaki S, Iwabuchi KA, Chantzoura E, Ruetz T, Johnsson A, et al. High-resolution analysis with novel cell-surface markers identifies routes to iPS cells. Nature. 2013;499:88–91.
Huang Y, Zhang H, Wang L, Tang C, Qin X, Wu X, et al. JMJD3 acts in tandem with KLF4 to facilitate reprogramming to pluripotency. Nat Commun. 2020;11:5061.
Zhuang Q, Li W, Benda C, Huang Z, Ahmed T, Liu P, et al. NCoR/SMRT co-repressors cooperate with c-MYC to create an epigenetic barrier to somatic cell reprogramming. Nat Cell Biol. 2018;20:400–12.
Seeburg PH, Colby WW, Capon DJ, Goeddel DV, Levinson AD. Biological properties of human c-Ha-ras1 genes mutated at codon 12. Nature. 1984;312:71–5.
Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, et al. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–5.
Luo Z, Qing X, Benda C, Huang Z, Zhang M, Huang Y, et al. Nuclear-cytoplasmic shuttling of class IIa histone deacetylases regulates somatic cell reprogramming. Cell Regen. 2019;8:21–9.
Chunduri NK, Storchova Z. The diverse consequences of aneuploidy. Nat Cell Biol. 2019;21:54–62.
Shahbazi MN, Wang T, Tao X, Weatherbee BAT, Sun L, Zhan Y, et al. Developmental potential of aneuploid human embryos cultured beyond implantation. Nat Commun. 2020;11:3987.
Yang Y, Shi L, Fu X, Ma G, Yang Z, Li Y, et al. Metabolic and epigenetic dysfunctions underlie the arrest of in vitro fertilized human embryos in a senescent-like state. PLoS Biol. 2022;20:e3001682.
Taapken SM, Nisler BS, Newton MA, Sampsell-Barron TL, Leonhard KA, McIntire EM, et al. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–4.
Gaztelumendi N, Nogues C. Chromosome instability in mouse embryonic stem cells. Sci Rep. 2014;4:5324.
Hutchins AP, Yang Z, Li Y, He F, Fu X, Wang X, et al. Models of global gene expression define major domains of cell type and tissue identity. Nucleic Acids Res. 2017;45:2354–67.
Steffens S, Fu X, He F, Li Y, Babarinde IA, Hutchins AP. DPre: computational identification of differentiation bias and genes underlying cell type conversions. Bioinformatics. 2020;36:1637–9.
Kareta MS, Gorges LL, Hafeez S, Benayoun BA, Marro S, Zmoos AF, et al. Inhibition of pluripotency networks by the Rb tumor suppressor restricts reprogramming and tumorigenesis. Cell Stem Cell. 2015;16:39–50.
Tapia N, Scholer HR. p53 connects tumorigenesis and reprogramming to pluripotency. J Exp Med. 2010;207:2045–8.
Kawamura T, Suzuki J, Wang YV, Menendez S, Morera LB, Raya A, et al. Linking the p53 tumour suppressor pathway to somatic cell reprogramming. Nature. 2009;460:1140–4.
Marion RM, Strati K, Li H, Murga M, Blanco R, Ortega S, et al. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–53.
Utikal J, Polo JM, Stadtfeld M, Maherali N, Kulalert W, Walsh RM, et al. Immortalization eliminates a roadblock during cellular reprogramming into iPS cells. Nature. 2009;460:1145–8.
Samavarchi-Tehrani P, Golipour A, David L, Sung HK, Beyer TA, Datti A, et al. Functional genomics reveals a BMP-driven mesenchymal-to-epithelial transition in the initiation of somatic cell reprogramming. Cell Stem Cell. 2010;7:64–77.
Li R, Liang J, Ni S, Zhou T, Qing X, Li H, et al. A mesenchymal-to-epithelial transition initiates and is required for the nuclear reprogramming of mouse fibroblasts. Cell Stem Cell. 2010;7:51–63.
Silva J, Barrandon O, Nichols J, Kawaguchi J, Theunissen TW, Smith A. Promotion of reprogramming to ground state pluripotency by signal inhibition. PLoS Biol. 2008;6:e253.
Medvedeva YA, Lennartsson A, Ehsani R, Kulakovskiy IV, Vorontsov IE, Panahandeh P, et al. EpiFactors: a comprehensive database of human epigenetic factors and complexes. Database. 2015;2015:bav067.
He J, Fu X, Zhang M, He F, Li W, Abdul MM, et al. Transposable elements are regulated by context-specific patterns of chromatin marks in mouse embryonic stem cells. Nat Commun. 2019;10:34.
Liang G, Taranova O, **a K, Zhang Y. Butyrate promotes induced pluripotent stem cell generation. J Biol Chem. 2010;285:25516–21.
Huangfu D, Maehr R, Guo W, Eijkelenboom A, Snitow M, Chen AE, et al. Induction of pluripotent stem cells by defined factors is greatly improved by small-molecule compounds. Nat Biotechnol. 2008;26:795–7.
Li D, Liu J, Yang X, Zhou C, Guo J, Wu C, et al. Chromatin accessibility dynamics during iPSC reprogramming. Cell Stem Cell. 2017;21:819–33.e6.
Suknuntha K, Ishii Y, Tao L, Hu K, McIntosh BE, Yang D, et al. Discovery of survival factor for primitive chronic myeloid leukemia cells using induced pluripotent stem cells. Stem Cell Res. 2015;15:678–93.
Kim J, Zaret KS. Reprogramming of human cancer cells to pluripotency for models of cancer progression. EMBO J. 2015;34:739–47.
Ferreiros A, Pedrosa P, Da Silva-Alvarez S, Triana-Martinez F, Vilas JM, Picallos-Rabina P, et al. Context-dependent impact of RAS oncogene expression on cellular reprogramming to pluripotency. Stem Cell Rep. 2019;12:1099–112.
Ito K, Nagata K, Ohta S, Matsuda Y, Ukai T, Yasuda I, et al. The oncogene-dependent resistance to reprogramming unveils cancer therapeutic targets. Cell Rep. 2022;39:110721.
Kong Y, Gimple RC, McVicar RN, Hodges AP, Yin J, Liu Y, et al. "Reprogram enablement" as an assay for identifying early oncogenic pathways by their ability to allow neoplastic cells to reacquire an epiblast state. Stem Cell Rep. 2020;15:761–75.
Ying QL, Wray J, Nichols J, Batlle-Morera L, Doble B, Woodgett J, et al. The ground state of embryonic stem cell self-renewal. Nature. 2008;453:519–23.
Shi Y, Do JT, Desponts C, Hahm HS, Scholer HR, Ding S. A combined chemical and genetic approach for the generation of induced pluripotent stem cells. Cell Stem Cell. 2008;2:525–8.
Malta TM, Sokolov A, Gentles AJ, Burzykowski T, Poisson L, Weinstein JN, et al. Machine learning identifies stemness features associated with oncogenic dedifferentiation. Cell. 2018;173:338–54.e15.
Warlich E, Kuehle J, Cantz T, Brugman MH, Maetzig T, Galla M, et al. Lentiviral vector design and imaging approaches to visualize the early stages of cellular reprogramming. Mol Ther. 2011;19:782–9.
He J, Babarinde IA, Sun L, Xu S, Chen R, Shi J, et al. Identifying transposable element expression dynamics and heterogeneity during development at the single-cell level with a processing pipeline scTE. Nat Commun. 2021;12:1456.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Korotkevich G, Sukhov V, Budin N, Shpak B, Artyomov MN, Sergushichev A. Fast gene set enrichment analysis. bioRxiv. 2021:060012.
Young MD, Wakefield MJ, Smyth GK, Oshlack A. Gene ontology analysis for RNA-seq: accounting for selection bias. Genome Biol. 2010;11:R14.
Hutchins AP, Jauch R, Dyla M, Miranda-Saavedra D. glbase: a framework for combining, analyzing and displaying heterogeneous genomic and high-throughput sequencing data. Cell Regeneration. 2014;3:1.
Buenrostro JD, Giresi PG, Zaba LC, Chang HY, Greenleaf WJ. Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat Methods. 2013;10:1213–8.
Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–9.
Zhang Y, Liu T, Meyer CA, Eeckhoute J, Johnson DS, Bernstein BE, et al. Model-based analysis of ChIP-Seq (MACS). Genome Biol. 2008;9:R137.
Ma G, Babarinde IA, Zhuang Q, Hutchins AP. Unified analysis of multiple ChIP-seq datasets. Methods Mol Biol. 2021;2198:451–65.
Heinz S, Benner C, Spann N, Bertolino E, Lin YC, Laslo P, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38:576–89.
Malik V, Glaser LV, Zimmer D, Velychko S, Weng M, Holzner M, et al. Pluripotency reprogramming by competent and incompetent POU factors uncovers temporal dependency for Oct4 and Sox2. Nat Commun. 2019;10:3477.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32150710521 and 32270597), and the Shenzhen Innovation Committee of Science and Technology (JCYJ20200109141018712). R.J. is supported by the Health and Medical Research Fund (06174006, 08192886), Research Grants Council of Hong Kong General Research Fund (RGC/GRF, Grant. No. 17128918, 17101120 and 17106622), and Collaborative Research Fund (CRF, Grant No. C7064-22G) and the Innovation Technology Commission Funding Scheme Health@InnoHK. Additional support was rendered by the Center for Computational Science and Engineering of the Southern University of Science and Technology.
Author information
Authors and Affiliations
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Fu, X., Zhuang, Q., Babarinde, I.A. et al. Restricting epigenetic activity promotes the reprogramming of transformed cells to pluripotency in a line-specific manner. Cell Death Discov. 9, 245 (2023). https://doi.org/10.1038/s41420-023-01533-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41420-023-01533-8
- Springer Nature Limited