Abstract
Reactive astrocytes (RA) secrete lipocalin-2 (LCN2) glycoprotein that regulates diverse cellular processes including cell death/survival, inflammation, iron delivery and cell differentiation. Elevated levels of LCN2 are considered as a biomarker of brain injury, however, the underlying regulatory mechanisms of its expression and release are not well understood. In this study, we investigated the role of astrocytic Na+/H+ exchanger 1 (NHE1) in regulating reactive astrocyte LCN2 secretion and neurodegeneration after stroke. Astrocyte specific deletion of Nhe1 in Gfap-CreER+/−;Nhe1f/f mice reduced astrogliosis and astrocytic LCN2 and GFAP expression, which was associated with reduced loss of NeuN+ and GRP78+ neurons in stroke brains. In vitro ischemia in astrocyte cultures triggered a significant increase of secreted LCN2 in astrocytic exosomes, which caused neuronal cell death and neurodegeneration. Inhibition of NHE1 activity during in vitro ischemia with its potent inhibitor HOE642 significantly reduced astrocytic LCN2+ exosome secretion. In elucidating the cellular mechanisms, we found that stroke triggered activation of NADPH oxidase (NOX)-NF-κB signaling and ROS-mediated LCN2 expression. Inhibition of astrocytic NHE1 activity attenuated NOX signaling and LCN2-mediated neuronal apoptosis and neurite degeneration. Our findings demonstrate for the first time that RA use NOX signaling to stimulate LCN2 expression and secretion. Blocking astrocytic NHE1 activity is beneficial to reduce LCN2-mediated neurotoxicity after stroke.
Similar content being viewed by others
Introduction
Reactive astrocytes (RA) in stroke and other neurodegenerative diseases often gain toxic functions and have negative impact on neuronal function and survival [1, 2]. For example, in response to acute ischemic stroke, RA release several proinflammatory cytokines (TNF-α, IL-6, and IFN-γ), free radicals, and matrix metalloproteinases (MMPs), which collectively leads to neuronal death, and contributes to infarct progression [3, 4]. Recent research shows that RA also increase the expression and release of LCN2 in response to infection, inflammation or injury [5, 6]. Increase in LCN2 protein level was detected in human post-mortem brain tissues of Alzheimer’s disease, Parkinson’s disease (PD), and multiple sclerosis patients [7,8,9]. Elevated plasma levels of LCN2 have also been detected in ischemic and hemorrhagic stroke patients and are associated with worsened clinical outcome [10] and LCN2 is currently employed as a biomarker of brain injury [11, 12]. LCN2 is shown to be involved in neuroinflammation and neuronal death in various animal brain injury models including cerebral ischemia [13, 14]. Enhanced LCN2 expression after cerebral ischemia contributes to neuronal death by promoting glial activation, neuroinflammation, and the BBB disruption [15]. LCN2 released from astrocytes induced caspase 3-mediated neuronal apoptosis in mice treated with psychotic stimulant methamphetamine (METH) [16]. Increased expression of LCN2 in RA in kainic acid-treated mice is associated with neuronal death due to induction of iron overload and oxidative stress [17].
Despite the significance of LCN2 in brain injury, the underlying regulatory mechanisms of its expression and release from astrocytes are not well understood. Endoplasmic reticulum (ER) stress has been shown to contribute to LCN2 mRNA and protein upregulation in astrocytes and renal cells [18, 19]. ROS-mediated ER stress activation in astrocytes leads to neuronal apoptosis in the METH-induced neuronal apoptosis model [16]. Several transcription factors have been identified as important regulators of LCN2 gene expression [5], including TNF-α and IL6-mediated activation of nuclear factor κB (NF-κB) and IκBζ [20]. However, how RA regulate LCN2 expression in ischemic stroke conditions remains to be established.
We recently reported that selective knockout of Nhe1 in astrocytes reduced reactive astrocyte formation and preserved the BBB integrity via stimulating Wnt/β-catenin signaling in stroke brains [21, 22]. In this study, we found that astrocyte-specific deletion of Nhe1 not only attenuated Lcn2 gene and protein expression in GFAP+ astrocytes but also reduced stroke-induced neurodegeneration. Activation of ER stress and UPR in astrocytes did not change LCN2 protein expression, instead, RA utilized NOX signaling to stimulate LCN2 expression and secretion. NHE1 protein activity in astrocytes is involved in accelerating NOX4-NF-κB signaling pathway. Thus, targeting astrocytic NHE1 protein is beneficial to reduce LCN2-mediated neurotoxicity after ischemic stroke.
Results
Nhe1 Astro-KO mice displayed enhanced resistance to neurodegeneration after stroke
Astrocyte specific knockout of Nhe1 in Nhe1 Astro-KO mice was established in our original study (Supplemental Fig. 1 of [21]), demonstrating with immunofluorescence analysis that the lack of NHE1 protein expression in GFAP+ astrocytes was observed in Nhe1 Astro-KO brains but not in the wild-type control brains. To investigate the impact of astrocyte-specific deletion of Nhe1 on neurodegeneration, wild-type control and Nhe1 Astro-KO mice underwent ischemic stroke (Fig. 1A). We first assessed neurodegeneration by examining loss of neuronal glucose-regulated protein 78 (GRP78), an ER chaperone protein important for ER and mitochondrial homeostasis [23], and loss of neuronal GRP78 protein is a marker for neurodegeneration [24]. As shown in Fig. 1B, wild-type brains expressed abundant levels of GRP78 in non-stroke brain regions. However, complete loss of GRP78 protein expression was detected in the ischemic core at 48 h reperfusion. Double immunofluorescence staining showed abundant expression of GRP78 in NeuN+ neurons in the non-stroke contralateral (CL) hemispheres (Fig. 1C). A concurrent loss of NeuN+/GRP78+ cells was detected in the peri-lesion of ipsilateral (IL) hemispheres (p < 0.0001). Interestingly, the stroke-induced loss of GRP78+/NeuN+ neurons was significantly reduced in the Nhe1 Astro-KO brains, compared to the wild-type control brains (p = 0.0178) (Fig. 1D, E), indicating less stroke-induced neurodegeneration in Nhe1 Astro-KO brains. These findings suggest that changes of astrocyte functions in the Nhe1 Astro-KO attenuated stroke-induced neurodegeneration.
A Experimental protocol and timeline. B Representative immunofluorescence images of GRP78 protein expression in wild-type and Nhe1 Astro-KO brains at 48 h Rp. High magnification images show peri nuclear GRP78 staining in peri-lesion areas indicated by white squares. C Representative confocal images of NeuN and GRP78 staining in neurons. Arrows: high GRP78 protein expression. Arrowheads: low GRP78 expression. D, E Summary data of GRP78+ and NeuN+ cells. Data are mean ± SD, n = 4. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 via one-way ANOVA. CL contralateral, IL Ipsilateral hemispheres.
Reduced astrocytic LCN2 protein expression in the Nhe1 Astro-KO mice was associated with preservation of GRP78+/NeuN+ neurons
In assessing RA in relation with degeneration of NeuN+ neurons, we found that loss of NeuN expressing neurons in the peri-lesion areas of wild-type brains was associated with reactive GFAP+ astrocytes expressing increased LCN2 protein (Arrows, Fig. 2A, B). An association between increased LCN2 protein expression and stroke-induced loss of GRP78 protein in neurons was also observed (Supplementary Fig. S1). In the ischemic peri-lesion areas of the wild-type stroke brains, RA exhibited both increased GFAP and LCN2 protein expression (Fig. 2D, E) that was not detected in neurons or microglia (Supplementary Fig. S2). In contrast, the Nhe1 Astro-KO brains exhibited less degeneration of NeuN+ neurons as well as less LCN2 upregulation in GFAP+ RA (Arrowheads, Fig. 2A, B), with a negative correlation between LCN2 expression in GFAP+ RA and NeuN+ neuronal counts (Fig. 2C, Pearson coefficient of r = −0.508, p = 0.022). These findings suggest a possible relationship between elevated astrocytic LCN2 expression and neurodegeneration in stroke brains. To further explore this possibility, we conducted analysis of Lcn2 mRNA expression in astrocytes isolated from wild-type and Nhe1 Astro-KO ischemic brains by qRT-PCR and bulk RNA sequencing dataset reported recently [22]. As shown in Fig. 2F, astrocytes from IL hemispheres of wild-type brains showed a significant increase in Lcn2 mRNA (p < 0.01). In contrast, Lcn2 mRNA in astrocytes from Nhe1 Astro-KO ischemic brains remained low (Fig. 2F). Taken together, these data strongly suggest that reduced LCN2 expression in GFAP+ RA in Nhe1 Astro-KO ischemic brains may play a role in preventing neuronal loss.
A Representative confocal images of NeuN and LCN2 expression in astrocytes at 48 h Rp. Arrows: high expression. Arrowheads: low expression. B Orthogonal sections from z stack confocal images showing association of NeuN+ neurons with LCN2+ astrocytes (arrows). C Negative correlation between LCN2 expression and NeuN+ intensity in wild-type and Nhe1 Astro-KO brains. r = −0.5079, p = 0.0222. D Representative confocal images of GFAP and LCN2 expression in astrocytes at 48 h Rp. Arrows: high expression. Arrowheads: low expression. E Summary data shows quantification of GFAP and LCN2 fluorescence signal intensity and summary data of GFAP and LCN2 double positive cells percentage. Data are mean ± SD, n = 5. *p < 0.05; **p < 0.01; ***p < 0.001; ****p < 0.0001 via one-way ANOVA and unpaired t-test. F (i) Cartoon illustrates isolation of astrocyte by MACS. (ii) Relative gene expression of LCN2 expressed as RPKM normalized values and RT-qPCR analysis of changes in expression of LCN2 mRNA in the astrocytes isolated from wild-type and Nhe1 Astro-KO brains at 24 h Rp. Data are mean ± SD n = 6, *p < 0.05 by unpaired t-test.
Astrocytic ER stress activation did not cause LCN2 protein upregulation
The mechanisms underlying LCN2 upregulation in RA are not well understood. Activation of PERK pathway, a main branch of the UPR [25] in astrocyte cultures by ER stressor thapsigargin (Tg) has been shown to increase expression of Lcn2 mRNA [18]. To determine whether ER stress-dependent mechanisms play a role in regulating astrocytic LCN2 expression in ischemic stroke, we first tested whether Tg differently induced ER stress responses and LCN2 expression in primary astrocyte cultutres isolated from wild-type (Nhe1+/+) and global Nhe1 KO (Nhe1−/−) mice. As shown in Fig. 3A (i) and (ii), Tg triggered significant upregulation of both ATF4 (p = 0.014) and GADD34 (p = 0.0057), two downstream proteins of PERK pathway, in Nhe1+/+ astrocyte cultutres but only GADD34 upregulation in global Nhe1−/− astrocytes (p = 0.0213). These changes were blocked by PERK inhibitor GSK2606414 (Fig. 3A). However, Tg treatment did not cause any significant changes of LCN2 protein expression in either Nhe1+/+ or global Nhe1−/− astrocytes (Fig. 3A (iii)). This was further illustrated by lack of ER stress induction in astrocytes after OGD/REOX (Supplementary Fig. S3), suggesting ER stress induction does not affect LCN2 expression in Nhe1+/+ or global Nhe1−/− astrocytes.
A Representative immunoblots showing the expression of ER stress proteins in astrocyte cultures from wild-type (+/+) and global Nhe1 KO (−/−) mice treated with ER stress inducer thapsigargin (Tg) 100 nM or the pathway inhibitor GSK2606414 (5 μM) for 24 h. (i-iii) Summary. Data are mean ± SD, n = 3. *p < 0.05; **p < 0.01 via one-way ANOVA. B, C are representative immunoblots showing the expression of ER stress proteins in brain homogenate and astrocytes isolated from wild-type and Nhe1 Astro-KO brain at 24 h Rp. CL Contralateral, IL Ipsilateral hemisphere. (i-iii) Quantification of p-eIF2α/t-eIF2α, ATF4 and CHOP protein bands. Data are expressed as relative change (fold) of the CL controls. Data are mean ± SD, n = 4. *p < 0.05; ** p < 0.01; *** p < 0.001; ****p < 0.0001 via one-way ANOVA.
We next examined whether ischemic stroke triggers PERK pathway activation differently in wild-type and Nhe1 Astro-KO brains. Immunoblotting analysis of p-eIF2α/t-eIF2α, ATF4 and CHOP protein expression in brain homogenates (Fig. 3B) revealed that ischemic stroke triggered a significant increase in the expression of ATF4 (p < 0.0001) and CHOP proteins (p = 0.002) in the wild-type ischemic brains but not in the Nhe1 Astro-KO ischemic brains (Fig. 3B (i) and (ii)). No significant changes of p-eIF2α/t-eIF2α expression were detected in either wild-type or Nhe1 Astro-KO ischemic brain homogenates (Fig. 3B (i)). Considering that our immunostaining data showed that loss of ER chaperone GRP78 was mainly in NeuN+ neurons but not in astrocytes, we speculate that the changes of ATF4 and CHOP expression in brain homogenates mainly represent neuronal changes. To validate this, MACS-isolated astrocytes were obtained from wild-type and Nhe1 Astro-KO brains. As shown in Fig. 3C, no changes in p-eIF2α/t-eIF2α expression were observed in wild-type or Nhe1 Astro-KO astrocytes. The expression of PERK pathway proteins ATF4 and CHOP in astrocytes was not detectable, which is consistent with other reports in mild ER stress [26]. Our results further suggest that stroke-induced astrocytic LCN2 upregulation is likely mediated by ER stress-independent mechanisms.
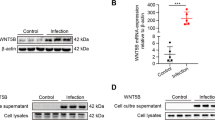
Ischemia induced increases in LCN2 secretion in astrocytic exosomes
RA communicate with neurons via secreting exosomes containing diverse signaling molecules [27,28,29]. LCN2 secreted from RA contributes to neuronal death in response to brain injury caused by intracerebral hemorrhage or exposure to antipsychotic drugs [16, 21].
Focal ischemic stroke model
Transient focal cerebral ischemia was induced by intraluminal occlusion of the left middle cerebral artery (MCA), as previously described [60]. Briefly, under 1.5% isoflurane, the left common carotid artery (CCA) was exposed via a midline pre-tracheal incision and the external carotid artery (ECA) and the CCA were ligated. To occlude the MCA, a rubber silicon-coated monofilament suture (size 6–0, diameter 0.09–0.11 mm, length 13 mm; diameter with coating 0.21 ± 0.02 mm; coating length 5 mm) was inserted into the ECA and advanced along the internal carotid artery 8–9 mm from the bifurcation of the carotid artery. For reperfusion, the suture was gently withdrawn 60 min after occlusion. Cranial and body temperatures were monitored with tympanic membrane and rectal temperature probes and maintained at 36.5 ± 0.5 oC throughout the experiment by a heating blanket.
Immunofluorescence staining and image analysis
Mouse brain sections
Mice were anesthetized and transcardially perfused as previously described [21]. Coronal brains sections (25 µm, at +1.1 mm bregma) were washed with PBS and incubated with blocking solution (10% normal goat serum (NGS), 5% BSA and 0.5% Triton X-100 in 0.01 M PBS) for 1 h at room temperature (RT) followed by incubation with primary antibodies (Supplementary Table 1). All the primary antibodies were diluted in the blocking solution containing 3% NGS and 0.3% Triton X-100 in PBS and incubated with brain sections overnight at 4 °C. On the following day, the sections or cultured cells were washed with PBS and then incubated with respective secondary Alexa 488/546 conjugated IgGs (1:200, Thermo scientific Life Technologies Corporation). DAPI (1:1000 in blocking solution) was used to stain the nucleus. For negative controls, brain sections were stained with the secondary antibody only. Fluorescent images were captured with either Olympus IX81 inverted microscope with a FV1000 laser scanning confocal system or NIKON A1R confocal microscope using a 40x, 60x or 100x, oil-immersion objective and 1024 × 1024 pixel resolution (0.103 μm/pixel).
Neuronal or astrocyte cultures
Cultutred cells grown on coverslips were fixed in 4% paraformaldehyde in PBS for 15 min. After rinsing, cells were incubated with a blocking solution (10% normal goat serum (NGS), 5% BSA and 0.5% Triton X-100 in 0.01 M PBS) for 1 h at RT followed by incubation with primary antibodies overnight at 4 °C. (Supplementary Table 1). After rinsing in PBS, cells were incubated with appropriate goat Alexa fluor 546/488 secondary antibodies (1:200, Thermo scientific Life Technologies Corporation) for 1 h at RT. Fluorescence images were captured with NIKON A1R confocal microscope using a 40x or 60x oil-immersion objective. For negative controls, cultures were stained with secondary antibodies only (Supplementary Fig. S7). Fluorescence images were quantified with either NIS elements (NIKON) or Fiji (NIH) software.
Immunoblotting
Brain tissue, isolated astrocytes, astrocyte cultures, astrocyte conditioned media (ACM) or exosomes were homogenized in RIPA lysis buffer (Thermo Scientific, Rockford, IL, USA) containing protease and phosphatase inhibitor mixture (Roche) and centrifuged at 4 °C for 30 min at 14,000 × g. The supernatant was transferred to a fresh tube and concentration was determined by BCA kit (Thermo Scientific). An equal amount of proteins were separated by 4–15% SDS-PAGE and electrotransferred onto a PVDF membrane as described before [61]. For exosomes, equal volume of protein was separated using 4–15% SDS-PAGE. After blocking, the membrane was incubated at 4 °C overnight with primary antibodies (Supplementary Table 1). After 1 h incubation at room temperature with an appropriate secondary antibody conjugated with horseradish peroxidase (anti-rabbit antibody or anti-mouse antibody, 1:1000–1:2000), protein bands were visualized using enhanced chemiluminescence agents. The densities of bands were measured with ImageJ. Changes of target protein expression were first normalized with internal loading control protein expression.
Isolation of astrocyte from mouse stroke brains by magnetic-activated cell sorting (MACS)
Single-cell suspension of astrocytes from the contralateral (CL) and ipsilateral (IL) hemispheres from wild-type and Nhe1 Astro-KO brains were prepared using Adult Brain Dissociation Kit (Miltenyi Biotec, Germany) following manufacturers’ protocol. At 24 h post-tMCAO, brains were removed and placed in d-PBS (ice-cold, pH 7.2). Cortical and striatum tissues were rapidly dissected on ice and separated in enzyme mixture with gentleMACS Octo Dissociator for 30 min at 37 °C. Digested tissues were dissociated into single-cell suspension by filtration through 70 μm MACS SmartStrainer and a series of centrifugation steps as per manufacturers protocol. A total of ∼2 × 106 cells were used for isolation of astrocytes by magnetic bead separation using anti-ASCA-2 microbead kit. The cell suspension was applied to the MS column, which was washed with 1−2 ml of PBS and the eluted fraction was pelleted at 300 × g for 10 min. The pelleted cells were further processed either for immunoblotting or RT-PCR analysis.
RT-qPCR analysis
RNA of MACS-isolated astrocytes was extracted using the Direct-zol™ RNA MicroPrep kit (Zymo Research, R2060). RNA quantification was carried out by measuring absorbance with spectrophotometer ND-1000 (NanoDrop). Reverse transcription was performed using the iScript Reverse Transcription Supermix (Bio-Rad, 1708840) according to the manufacturer’s protocol. All RNA isolated from cell pellets was converted into cDNA. Quantitative RT-PCR was performed using iTaq Universal SYBR Green Supermix (Bio-Rad, 172-5120) on a CFX 96 Touch Real-Time PCR Detection System. All relative gene expression analyses were performed using the 2 − ΔΔCt method in a minimum of four animals per group with duplicate reactions for each gene evaluated. Following primer sequences were used as follows.
Gene | Forward (5’ →3’) | Reverse (5’ →3’) |
|---|---|---|
GAPDH | AACTTTGGCATTGTGGAAGG | ACACATTGGGGGTAGGAACA |
LCN2 | TGGCCCTGAGTGTCATGTG | CTCTTGTAGCTCATAGATGGTGC |
Primary cortical astrocyte or neuronal cultures
Dissociated cortical astrocyte cultures were established as described before [62]. Cerebral cortices were removed from P1-3 C57BL/6 J mice. The cortices were digested with TrypLE™ Express Enzyme (Gibco) for 15 min at 37 °C. The dissociated cells were rinsed and resuspended in DMEM (Gibco) containing 10% fetal bovine serum and 1% antibiotic-antimycotic (Gibco). Subsequently, the viable cells (2 × 106 cells) were plated on 75 cm2 flasks. Cultures were maintained in a 5% CO2 atmosphere at 37 °C and refed every 3 d throughout the study. Upon reaching confluency, astrocytes were seeded into 60 mm dishes at a density of 106 cells per plate. And 10–14 day in cultures (DIV) were used for study.
Primary cortical neuronal cultures were established from embryonic days 14–16 mouse embryos (C57BL/6 J) as described previously [63]. Briefly, cortices were dissected in the ice-cold Hanks balanced salt solution. The tissue was treated with 0.25 mg/ml trypsin at 37 °C for 20 min. The cells were centrifuged at 1200 RPM at RT for 5 mins. The cell suspension was diluted in B-27 supplemented neurobasal medium containing 25 μM L-glutamine (Thermo Scientific) and 1% antibiotic-antimycotic (Gibco). The cells (1000 cells/mm2) were seeded on glass coverslips coated with poly-d-Lysine. Cultures were incubated at 37 °C in an incubator with 5% CO2 and atmospheric air. Cultures were fed every 3 days with fresh media and used for experiments at 7 DIV.
In vitro ischemia of astrocyte cultures and drug treatment
In vitro ischemia was mimicked with well-established oxygen and glucose deprivation/reoxygenation (OGD/R) model as described previously [64]. Astrocyte cultures were rinsed twice with an isotonic OGD solution containing (in mM, pH 7.4): 0 glucose, 20 NaHCO3, 120 NaCl, 5.36 KCl, 0.33 Na2HPO4, 0.44 KH2PO4, 1.27 CaCl2, 0.81 MgSO4. Cells were incubated in OGD solution in a hypoxic incubator at 37oC (Forma model 3130, Thermo Forma, Marietta, OH) equilibrated with 95% N2 and 5% CO2 for 3 h. Normoxic control cells were incubated in 5% CO2 and atmospheric air in isotonic control buffer containing 5.5 mM glucose with the rest of the components in the buffer identical to the isotonic OGD solution. For reoxygenation (R), OGD-treated cells were incubated with DMEM and incubated at 37oC in 5% CO2 and atmospheric air.
In some experiments, astrocyte cultures were treated with ER stress inducer thapsigargin (100 nM) alone or in combination with GSK2606414 (5 μM), a potent inhibitor of PERK kinase signaling [18], or NHE1 inhibitor HOE642 (1 μM) for 24 h. To inhibit NOX activity, astrocytes were treated with general NOX inhibitor DPI or NOX4 specific inhibitor GKT137831 during OGD 3 h and Reox 1 h. We evaluated the dose-dependent effects of Re-LCN2 protein (1, 2, 4, and 6 µg/ml) on neuronal death and neurite degeneration of neuronal cultures. Re-LCN2 at a concentration of 2 µg/ml induced a significant increase in PI+ neurons but without increase in neurite degeneration (data not shown). A higher concentration of Re-LCN2 (4 µg/ml) significantly increased both neuronal death as well as neurite degeneration (Supplementary Fig. S6), which is consistent with previously published reports [16].
Exosome isolation from ACM by Size Exclusion Chromatography (SEC)
Exosomes were isolated from astrocyte conditioned medium (ACM) obtained from astrocyte cultures at 10–14 DIV (4 × 106cells/15 cm dish). ACM was centrifuged at 300 × g for 10 min at 4 °C to remove cell debris, followed by centrifugation at 2000 × g for 60 min at 4 °C and the supernatant was collected. The supernatant was further concentrated through Centricon Plus-70 Centrifugal Filter (Sigma, MO, USA) at 3500 × g for 30 min at 4 °C. The concentrate was filtered through a 0.22 μm filter (Celltreat Scientific Products, MA, USA) to remove contaminating apoptotic bodies and microvesicles, and was overlaid on qEV SEC columns (Izon Science, MA, USA) followed by elution with PBS. After discarding the void volume (~2.85 ml), six 500 µl fractions were collected following the manufacturer’s instruction and then each fraction was analyzed by nanoparticle tracking analysis (NTA) to pool EV-enriched fractions (f 1-3), while the pooled fractions 4–6 was non-EV fraction. The EV-enriched fraction (f 1-3) was further concentrated with Amicon Ultra-4 Centrifugal Filter Unit (Millipore, MA, USA) to obtain a final EV volume of ~60 µl.
Isolation of exosomes from ischemic mouse brains
Exosomes were isolated from mouse brains as described previously with slight modifications [1]. Briefly, fresh frozen brains (CL and IL hemispheres) from wild-type and Nhe1 Astro-KO mice at 24 h after ischemic stroke were sliced with a razor blade (on ice) to generate 2–3 mm3 sections. The sections were dissociated in the enzyme mixture from the neural dissociation kit (Miltenyi Biotec, Germany) and incubated at 37 °C for 20 min. After incubation, the samples were triturated slowly using a fire-polished pasteur pipette and the suspension was centrifuged at 300 g for 5 min at 4 °C. The pellet was collected (nominated as “total brain homogenate”) in lysis buffer (with protease and phosphatase inhibitors (PI/PS)), homogenized and stored at −80 °C for later use. The supernatant was transferred to a 15 ml-tube and then centrifuged at 2000 g for 10 min at 4 °C. This supernatant was then transferred to an eppendorf tube and then centrifuged at 10,000 g for 30 min at 4 °C. The supernatant was filtered through a 0.22 μm pore-size filter (Millipore) and overlaid on three sucrose cushions (2 ml each of 2.5 M, 1.3 M, and 0.6 M in 20 mM HEPES), then ultracentrifuged at 180,000 g for 3 h at 4 °C (Optima-XE SW41 Beckman Coulter) using SW41Ti rotor (Beckman). After the spin the top 1 ml of the gradient was discarded. Three 2 ml fractions were subsequently collected from the top to the bottom and the last 1 ml at the bottom of the tube was discarded. Each fraction was diluted with ice-cold PBS and spun at 100,000 × g at 4 °C (SW41Ti Beckman) for 1 h to pellet the vesicles. Following spin completion, the supernatant was discarded, and pellets were collected in ice-cold lysis buffer (with PI/PS). The vesicles in fractions 1–3 was characterized by NTA and TEM analysis to determine the nature of the vesicles and exosomes. The extracellular exosome vesicles ranging from 30 to 140 nm were identified in fraction 2 and equal quantities of exosome volume were used for immunoblotting.
Nanoparticle tracking analysis (NTA)
Exosome concentration and size distribution were characterized by NTA with a NanoSight NS300 instrument (Malvern, Worcestershire, UK) and corresponding software version NTA3.4, Exosomes were diluted (1:100) in filtered sterile ddH2O to a final volume of 1 ml to achieve a concentration within 107–108 range for optimal analysis. Three videos of 30 s were captured under the following conditions: cell temperature: 22 °C; Syringe speed: 20 µl/s, camera level: 15. The mean size and exosome concentration (particles/ml) were calculated by integrating the data from three records adjusting a detecting threshold of 5.
Transmission electron microscopy (TEM)
The isolated exosomes were centrifuged using an airfuge at 100,000 × g with 25 PSI for 45 min. The pellet was fixed with cold 2.5% v/v glutaraldehyde in 0.1 M PBS, rinsed in PBS, dehydrated through a graded series of ethanol and embedded in Epon. Ultra-thin sections (65 nm) were stained with uranyl acetate and Reynold’s lead citrate. A JEOL 1400 TEM was used for imaging at the Center for Biologic Imaging at the University of Pittsburgh.
Neuronal viability assay
Neuronal viability in neuronal cultures was assessed by propidium iodide (PI) uptake and retention of calcein. Briefly, cultured neurons were rinsed with the HEPES-MEM buffer and incubated with 1 μg/ml calcein-AM and 10 μg/ml PI in the same buffer at 37 °C for 35 min. Cells were then rinsed with HEPES-MEM buffer and visualized using a Nikon TiE inverted epifluorescence microscope equipped with a 20× objective lens. Calcein or PI fluorescence was captured using ANDOR camera. Cells from each treatment group were counted and the viability was expressed as the ratio of calcein-positive cells to the sum of calcein-positive and PI-positive cells.
Neuronal apoptosis and neurite degeneration
Neuronal cultures were fixed in 4% PFA and immunostaining was performed using rabbit anti-cleaved caspase-3 (1:100) and chicken anti-MAP-2 (1:2000) antibodies. Confocal images (3–5 images/ experimental condition) were acquired using NIKON A1R confocal microscope. The number of cells expressing the cleaved caspase-3 were counted and % of apoptosis was expressed as a ratio of caspase-3+/DAPI+ cells. The neurite degeneration was quantified by counting the number of MAP-2 expressing neurites showing focal swellings.
Measurement of NADPH oxidase activity
NADPH oxidase activity assay was modified from Tarpey et al. [41]. Primary astrocytes treated with different conditions were washed twice with ice-cold PBS 1× solution (137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4 and 1.8 mM KH2PO4), and then scraped from the petri dish and resuspended in 250 μl PBS. For the chemiluminescence assay, lucigenin (250 μM) and NADPH (150 μM) were added and incubated with 100 μl of samples with or without SOD (15 IU) and the luminescence was recorded using a luminometer every 15 mins (PerkinElmer, Victor NivoTM). The NADPH induced the highest luminescence value minus the basal value was the NADPH oxidase activity. The value was expressed as Relative Light Units (RLU)/min.
ROS measurement
ROS measurement was performed using the nonfluorescent cell-permeant compound 2’-7’-dichlorofluorescein diacetate (DCFH-DA) according to manufacturer’s instructions. DCFH-DA is hydrolyzed by intracellular esterases to dichlorofluorescin (DCFH) which is trapped within the cell. This nonfluorescent molecule is then oxidized to fluorescent dichlorofluorescin by action of cellular oxidants. Primary astrocytes treated with different conditions were washed twice with PBS 1× solution and cells were loaded with 5 μM DCFH-DA diluted in serum free media for 30 min at 37 °C. Cells were washed three times with PBS and fluorescent images were captured using a Nikon TiE inverted epifluorescence microscope equipped with a 20× objective lens.
Histochemistry for detection of iron
For the detection of non-heme (mostly Fe3+ form) iron the DAB-enhanced Perl’s iron stain was used, as described previously with some modifications [65]. Briefly, brain sections were mounted on glass slides and allowed to dry overnight. The slides were washed in 0.01 M PBS and endogenous peroxidases were blocked by incubation with 0.3% H2O2 for 30 min. Subsequently, sections were washed again in PBS, and Perl’s solution (1:1 mix of 5% potassium ferrocyanide and 5% HCL both prepared in milliQ H2O) was prepared shortly before use. Sections were then incubated in Perl’s solution for 12 h and washed in milliQ H2O for 15 min. Diaminobenzidine (DAB) reaction was performed by incubating the sections with DAB/H2O2 solution (Vector labs) for 30 min followed by quick washing with PBS. The sections were dehydrated and coverslipped using DPX mounting media. Iron staining was performed in dark as much as possible.
Statistical Analysis
A total of 60 mice (male and female, 3–4 months old) were used in the study. Sample sizes were determined based on pilot studies or our previously published work. All mice were randomly assigned to the procedures including both male and females in each group. All data analyses were performed by the investigators blinded to mouse genotypes and/or experimental group assignments. Mean values were reported together with the standard deviation (SD). Student’s two-tailed t test was used for comparison of two experimental groups. For three or more groups, multiple comparisons were done using one-way ANOVA where appropriate followed by appropriate comparisons test for multiple pairwise examinations. Changes were identified as statisticaly significant if p values were less than 0.05. Data was graphed using GraphPad Prism, version 8 (GraphPad Software, Inc., San Diego, CA) or Sigma plot (Systat Software Inc. San Jose, CA).
Data availability
Data can be made available upon valid request.
References
Chun H, Im H, Kang YJ, Kim Y, Shin JH, Won W, et al. Severe reactive astrocytes precipitate pathological hallmarks of Alzheimer’s disease via H(2)O(2)(-) production. Nat Neurosci. 2020;23:1555–66.
Patabendige A, Singh A, Jenkins S, Sen J, Chen R. Astrocyte Activation in Neurovascular Damage and Repair Following Ischaemic Stroke. Int J Mol Sci. 2021;22:4280.
Liu Z, Chopp M. Astrocytes, therapeutic targets for neuroprotection and neurorestoration in ischemic stroke. Prog Neurobiol. 2016;144:103–20.
Buffo A, Rolando C, Ceruti S. Astrocytes in the damaged brain: molecular and cellular insights into their reactive response and healing potential. Biochem Pharm. 2010;79:77–89.
Suk K. Lipocalin-2 as a therapeutic target for brain injury: An astrocentric perspective. Prog Neurobiol. 2016;144:158–72.
**ng C, Wang X, Cheng C, Montaner J, Mandeville E, Leung W, et al. Neuronal production of lipocalin-2 as a help-me signal for glial activation. Stroke. 2014;45:2085–92.
Kim BW, Jeong KH, Kim JH, ** M, Kim JH, Lee MG, et al. Pathogenic Upregulation of Glial Lipocalin-2 in the Parkinsonian Dopaminergic System. J Neurosci. 2016;36:5608–22.
Al Nimer F, Elliott C, Bergman J, Khademi M, Dring AM, Aeinehband S, et al. Lipocalin-2 is increased in progressive multiple sclerosis and inhibits remyelination. Neurol Neuroimmunol Neuroinflamm. 2016;3:e191.
Dekens DW, Naudé PJ, Engelborghs S, Vermeiren Y, Van Dam D, Oude, Voshaar RC, et al. Neutrophil Gelatinase-Associated Lipocalin and its Receptors in Alzheimer’s Disease (AD) Brain Regions: Differential Findings in AD with and without Depression. J Alzheimers Dis. 2017;55:763–76.
Hochmeister S, Engel O, Adzemovic MZ, Pekar T, Kendlbacher P, Zeitelhofer M, et al. Lipocalin-2 as an Infection-Related Biomarker to Predict Clinical Outcome in Ischemic Stroke. PLoS One. 2016;11:e0154797.
Jha MK, Suk K. Glia-based biomarkers and their functional role in the CNS. Expert Rev Proteom. 2013;10:43–63.
Chan CP, Jiang HL, Leung LY, Wan WM, Cheng NM, Ip WS, et al. Multiple atherosclerosis-related biomarkers associated with short- and long-term mortality after stroke. Clin Biochem. 2012;45:1308–15.
** M, Jang E, Suk K. Lipocalin-2 Acts as a Neuroinflammatogen in Lipopolysaccharide-injected Mice. Exp Neurobiol. 2014;23:155–62.
Wang G, Weng YC, Han X, Whaley JD, McCrae KR, Chou WH. Lipocalin-2 released in response to cerebral ischaemia mediates reperfusion injury in mice. J Cell Mol Med. 2015;19:1637–45.
** M, Kim JH, Jang E, Lee YM, Soo Han H, Woo DK, et al. Lipocalin-2 deficiency attenuates neuroinflammation and brain injury after transient middle cerebral artery occlusion in mice. J Cereb Blood Flow Metab. 2014;34:1306–14.
Chen X, Qiu F, Zhao X, Lu J, Tan X, Xu J, et al. Astrocyte-Derived Lipocalin-2 Is Involved in Mitochondrion-Related Neuronal Apoptosis Induced by Methamphetamine. ACS Chem Neurosci. 2020;11:1102–16.
Shin HJ, Jeong EA, Lee JY, An HS, Jang HM, Ahn YJ. et al. Lipocalin-2 Deficiency Reduces Oxidative Stress and Neuroinflammation and Results in Attenuation of Kainic Acid-Induced Hippocampal Cell Death. Antioxidants (Basel). 2021;10:100
Smith HL, Freeman OJ, Butcher AJ, Holmqvist S, Humoud I, Schätzl T, et al. Astrocyte Unfolded Protein Response Induces a Specific Reactivity State that Causes Non-Cell-Autonomous Neuronal Degeneration. Neuron. 2020;105:855–66.e5.
El Karoui K, Viau A, Dellis O, Bagattin A, Nguyen C, Baron W, et al. Endoplasmic reticulum stress drives proteinuria-induced kidney lesions via Lipocalin 2. Nat Commun. 2016;7:10330.
Zhao W, Bendickson L, Nilsen-Hamilton M. The Lipocalin2 Gene is Regulated in Mammary Epithelial Cells by NFκB and C/EBP In Response to Mycoplasma. Sci Rep. 2020;10:7641.
Begum G, Song S, Wang S, Zhao H, Bhuiyan MIH, Li E, et al. Selective knockout of astrocytic Na(+)/H(+) exchanger isoform 1 reduces astrogliosis, BBB damage, infarction, and improves neurological function after ischemic stroke. Glia. 2018;66:126–44.
Song S, Huang H, Guan X, Fiesler V, Bhuiyan MIH, Liu R, et al. Activation of endothelial Wnt/β-catenin signaling by protective astrocytes repairs BBB damage in ischemic stroke. Prog Neurobiol. 2021;199:101963.
Goldenberg-Cohen N, Raiter A, Gaydar V, Dratviman-Storobinsky O, Goldstein T, Weizman A, et al. Peptide-binding GRP78 protects neurons from hypoxia-induced apoptosis. Apoptosis. 2012;17:278–88.
Gorbatyuk MS, Gorbatyuk OS. The Molecular Chaperone GRP78/BiP as a Therapeutic Target for Neurodegenerative Disorders: A Mini Review. J Genet Syndr Gene Ther. 2013;4:128.
Nakka VP, Gusain A, Raghubir R. Endoplasmic Reticulum Stress Plays Critical Role in Brain Damage After Cerebral Ischemia/Reperfusion in Rats. Neurotox Res. 2009;17:189.
Wang Y, Chen Y, Zhou Q, Xu J, Qian Q, Ni P. et al. Mild Endoplasmic Reticulum Stress Protects Against Lipopolysaccharide-Induced Astrocytic Activation and Blood-Brain Barrier Hyperpermeability. Front Cell Neurosci. 2018;12:222.
Théry C, Witwer KW, Aikawa E, Alcaraz MJ, Anderson JD, Andriantsitohaina R, et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): a position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J Extracell Vesicles. 2018;7:1535750.
Liu W, Bai X, Zhang A, Huang J, Xu S, Zhang J. et al. Role of Exosomes in Central Nervous System Diseases. Frontiers in Molecular Neuroscience. 2019;12:240.
Gupta A, Pulliam L. Exosomes as mediators of neuroinflammation. J Neuroinflammation. 2014;11:68.
Dong M, ** G, Keep RF, Hua Y. Role of iron in brain lipocalin 2 upregulation after intracerebral hemorrhage in rats. Brain Res. 2013;1505:86–92.
Vella LJ, Scicluna BJ, Cheng L, Bawden EG, Masters CL, Ang CS, et al. A rigorous method to enrich for exosomes from brain tissue. J Extracell Vesicles. 2017;6:1348885.
Borkham-Kamphorst E, Van de Leur E, Meurer SK, Buhl EM, Weiskirchen R. N-Glycosylation of Lipocalin 2 Is Not Required for Secretion or Exosome Targeting. Front Pharm. 2018;9:426.
Batista BS, Eng WS, Pilobello KT, Hendricks-Muñoz KD, Mahal LK. Identification of a conserved glycan signature for microvesicles. J Proteome Res. 2011;10:4624–33.
Fujiwara Y, Tsuchiya H, Sakai N, Shibata K, Fujimura A, Koshimizu TA. Lipopolysaccharide-induced inflammation or unilateral ureteral obstruction yielded multiple types of glycosylated Lipocalin 2. J Inflamm (Lond). 2016;13:7.
Huh JW, Raghupathi R, Laurer HL, Helfaer MA, Saatman KE. Transient loss of microtubule-associated protein 2 immunoreactivity after moderate brain injury in mice. J Neurotrauma. 2003;20:975–84.
Makhezer N, Ben Khemis M, Liu D, Khichane Y, Marzaioli V, Tlili A, et al. NOX1-derived ROS drive the expression of Lipocalin-2 in colonic epithelial cells in inflammatory conditions. Mucosal Immunol. 2019;12:117–31.
Lam TI, Brennan-Minnella AM, Won SJ, Shen Y, Hefner C, Shi Y, et al. Intracellular pH reduction prevents excitotoxic and ischemic neuronal death by inhibiting NADPH oxidase. Proc Natl Acad Sci USA. 2013;110:E4362–8.
Liu Y, Kintner DB, Chanana V, Algharabli J, Chen X, Gao Y, et al. Activation of microglia depends on Na+/H+ exchange-mediated H+ homeostasis. J Neurosci. 2010;30:15210–20.
Cooney SJ, Bermudez-Sabogal SL, Byrnes KR. Cellular and temporal expression of NADPH oxidase (NOX) isotypes after brain injury. J Neuroinflammation. 2013;10:155.
Park MW, Cha HW, Kim J, Kim JH, Yang H, Yoon S, et al. NOX4 promotes ferroptosis of astrocytes by oxidative stress-induced lipid peroxidation via the impairment of mitochondrial metabolism in Alzheimer’s diseases. Redox Biol. 2021;41:101947.
Tarpey MM, White CR, Suarez E, Richardson G, Radi R, Freeman BA. Chemiluminescent detection of oxidants in vascular tissue. Lucigenin but not coelenterazine enhances superoxide formation. Circ Res. 1999;84:1203–11.
Lim D, Jeong JH, Song J. Lipocalin 2 regulates iron homeostasis, neuroinflammation, and insulin resistance in the brains of patients with dementia: Evidence from the current literature. CNS Neurosci Ther. 2021;27:883–94.
Escartin C, Galea E, Lakatos A, O’Callaghan JP, Petzold GC, Serrano-Pozo A, et al. Reactive astrocyte nomenclature, definitions, and future directions. Nat Neurosci. 2021;24:312–25.
Pekny M, Wilhelmsson U, Tatlisumak T, Pekna M. Astrocyte activation and reactive gliosis-A new target in stroke? Neurosci Lett. 2019;689:45–55.
Elicegui A, Schmitz T, Revuelta M. Astrogliosis after ischemic stroke: Neuroprotection or neuroinflammation. Am J Aging Sci Res. 2021;2:9–12.
Kim JH, Ko PW, Lee HW, Jeong JY, Lee MG, Kim JH, et al. Astrocyte-derived lipocalin-2 mediates hippocampal damage and cognitive deficits in experimental models of vascular dementia. Glia. 2017;65:1471–90.
Mesquita SD, Ferreira AC, Falcao AM, Sousa JC, Oliveira TG, Correia-Neves M, et al. Lipocalin 2 modulates the cellular response to amyloid beta. Cell Death Differ. 2014;21:1588–99.
Zhou Y, Tan J, Dai Y, Yu Y, Zhang Q, Meyerhoff ME. Synthesis and nitric oxide releasing properties of novel fluoro S-nitrosothiols. Chem Commun (Camb). 2019;55:401–4.
Flo TH, Smith KD, Sato S, Rodriguez DJ, Holmes MA, Strong RK, et al. Lipocalin 2 mediates an innate immune response to bacterial infection by sequestrating iron. Nature. 2004;432:917–21.
Devireddy LR, Gazin C, Zhu X, Green MR. A cell-surface receptor for lipocalin 24p3 selectively mediates apoptosis and iron uptake. Cell. 2005;123:1293–305.
Nielsen BS, Borregaard N, Bundgaard JR, Timshel S, Sehested M, Kjeldsen L. Induction of NGAL synthesis in epithelial cells of human colorectal neoplasia and inflammatory bowel diseases. Gut. 1996;38:414–20.
Kintner DB, Su G, Lenart B, Ballard AJ, Meyer JW, Ng LL, et al. Increased tolerance to oxygen and glucose deprivation in astrocytes from Na(+)/H(+) exchanger isoform 1 null mice. Am J Physiol Cell Physiol. 2004;287:C12–21.
Luo J, Sun D. Physiology and pathophysiology of Na(+)/H(+) exchange isoform 1 in the central nervous system. Curr Neurovasc Res. 2007;4:205–15.
Kalluri R, LeBleu VS. The biology, function, and biomedical applications of exosomes. Science. 2020;367:eaau6977.
Chalmin F, Ladoire S, Mignot G, Vincent J, Bruchard M, Remy-Martin JP, et al. Membrane-associated Hsp72 from tumor-derived exosomes mediates STAT3-dependent immunosuppressive function of mouse and human myeloid-derived suppressor cells. J Clin Invest. 2010;120:457–71.
Pironti G, Strachan RT, Abraham D. Mon-Wei Yu S, Chen M, Chen W, et al. Circulating Exosomes Induced by Cardiac Pressure Overload Contain Functional Angiotensin II Type 1 Receptors. Circulation. 2015;131:2120–30.
Savina A, Furlán M, Vidal M, Colombo MI. Exosome release is regulated by a calcium-dependent mechanism in K562 cells. J Biol Chem. 2003;278:20083–90.
Paroutis P, Touret N, Grinstein S. The pH of the secretory pathway: measurement, determinants, and regulation. Physiol (Bethesda). 2004;19:207–15.
Percie du Sert N, Hurst V, Ahluwalia A, Alam S, Avey MT, Baker M, et al. The ARRIVE guidelines 2.0: Updated guidelines for reporting animal research. PLoS Biol. 2020;18:e3000410.
Begum G, Yuan H, Kahle KT, Li L, Wang S, Shi Y, et al. Inhibition of WNK3 Kinase Signaling Reduces Brain Damage and Accelerates Neurological Recovery After Stroke. Stroke. 2015;46:1956–65.
Huang H, Bhuiyan MIH, Jiang T, Song S, Shankar S, Taheri T, et al. A Novel Na(+)-K(+)-Cl(-) Cotransporter 1 Inhibitor STS66* Reduces Brain Damage in Mice After Ischemic Stroke. Stroke. 2019;50:1021–5.
Su G, Kintner DB, Flagella M, Shull GE, Sun D. Astrocytes from Na(+)-K(+)-Cl(-) cotransporter-null mice exhibit absence of swelling and decrease in EAA release. Am J Physiol Cell Physiol. 2002;282:C1147–60.
Begum G, Yan HQ, Li L, Singh A, Dixon CE, Sun D. Docosahexaenoic acid reduces ER stress and abnormal protein accumulation and improves neuronal function following traumatic brain injury. J Neurosci. 2014;34:3743–55.
Kintner DB, Look A, Shull GE, Sun D. Stimulation of astrocyte Na+/H+ exchange activity in response to in vitro ischemia depends in part on activation of ERK1/2. Am J Physiol Cell Physiol. 2005;289:C934–45.
Dekens DW, Naudé PJW, Keijser JN, Boerema AS, De Deyn PP, Eisel ULM. Lipocalin 2 contributes to brain iron dysregulation but does not affect cognition, plaque load, and glial activation in the J20 Alzheimer mouse model. J Neuroinflammation. 2018;15:330.
Acknowledgements
This work was supported by United States National Institute of Health (NIH) grants, R01 NS110755-01A1 (G.B.), S10OD016236 (S.W.), R01NS118747 and R01NS125490 (Y.Y.).
Author information
Authors and Affiliations
Contributions
GB conceived and designed the study, and supervised the project with RL. RL, JW, YC, JC, OC, and MS performed the research; SJ, YY, SJ, TW, DS, collaborated; GB, and RL, analyzed the data; GB and RL wrote the paper.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing interests.
Ethical approval and consent to participate
Not applicable.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Edited by Professor Massimiliano Agostini
Rights and permissions
Open Access This article is licensed under a Creative Commons Attribution 4.0 International License, which permits use, sharing, adaptation, distribution and reproduction in any medium or format, as long as you give appropriate credit to the original author(s) and the source, provide a link to the Creative Commons license, and indicate if changes were made. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in a credit line to the material. If material is not included in the article’s Creative Commons license and your intended use is not permitted by statutory regulation or exceeds the permitted use, you will need to obtain permission directly from the copyright holder. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/.
About this article
Cite this article
Liu, R., Wang, J., Chen, Y. et al. NOX activation in reactive astrocytes regulates astrocytic LCN2 expression and neurodegeneration. Cell Death Dis 13, 371 (2022). https://doi.org/10.1038/s41419-022-04831-8
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41419-022-04831-8
- Springer Nature Limited
This article is cited by
-
The Transcription Factor Ets1 Influences Axonal Growth via Regulation of Lcn2
Molecular Neurobiology (2024)
-
The cGAS-STING-YY1 axis accelerates progression of neurodegeneration in a mouse model of Parkinson’s disease via LCN2-dependent astrocyte senescence
Cell Death & Differentiation (2023)
-
Scientific Landscape of Oxidative Stress in Stroke: From a Bibliometric Analysis to an in-Depth Review
Neurochemical Research (2023)
-
Dual Effect of Carnosine on ROS Formation in Rat Cultured Cortical Astrocytes
Molecular Neurobiology (2023)