Abstract
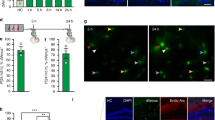
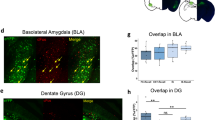
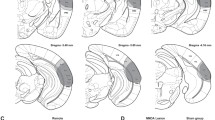
Contextual fear conditioning has been shown to activate a set of “fear ensemble” cells in the hippocampal dentate gyrus (DG) whose reactivation is necessary and sufficient for expression of contextual fear. We previously demonstrated that extinction learning suppresses reactivation of these fear ensemble cells and activates a competing set of DG cells—the “extinction ensemble.” Here, we tested whether extinction was sufficient to suppress reactivation in other regions and used single nucleus RNA sequencing (snRNA-seq) of cells in the dorsal dentate gyrus to examine how extinction affects the transcriptomic activity of fear ensemble and fear recall-activated cells. Our results confirm the suppressive effects of extinction in the dorsal and ventral dentate gyrus and demonstrate that this same effect extends to fear ensemble cells located in the dorsal CA1. Interestingly, the extinction-induced suppression of fear ensemble activity was not detected in ventral CA1. Our snRNA-seq analysis demonstrates that extinction training markedly changes transcription patterns in fear ensemble cells and that cells activated during recall of fear and recall of extinction have distinct transcriptomic profiles. Together, our results indicate that extinction training suppresses a broad portion of the fear ensemble in the hippocampus, and this suppression is accompanied by changes in the transcriptomes of fear ensemble cells and the emergence of a transcriptionally unique extinction ensemble.





Similar content being viewed by others
Data availability
Sequence data in this publication have been deposited in the National Center for Biotechnology Single Read Archive (SRA). Other data, including behavior, cell counts, metadata, and protocols/scripts are available on the Texas Data Repository.
References
Abramowitz JS, Deacon BJ, Whiteside SPH. Exposure therapy for anxiety. 2nd ed. Principles and practice. New York, NY: Guilford Press; 2019. p. 478.
Myers KM, Davis M. Behavioral and neural analysis of extinction. Neuron. 2002;36:567–84.
Rescorla RA. Spontaneous recovery. Learn Mem. 2004;11:501–9.
Heldt SA, Stanek L, Chhatwal JP, Ressler KJ. Hippocampus-specific deletion of BDNF in adult mice impairs spatial memory and extinction of aversive memories. Mol Psychiatry. 2007;12:656–70.
Bernier BE, Lacagnina AF, Ayoub A, Shue F, Zemelman BV, Krasne FB, et al. Dentate gyrus contributes to retrieval as well as encoding: evidence from context fear conditioning, recall, and extinction. J Neurosci. 2017;37:6359–71.
Lacagnina AF, Brockway ET, Crovetti CR, Shue F, McCarty MJ, Sattler KP, et al. Distinct hippocampal engrams control extinction and relapse of fear memory. Nat Neurosci. 2019;22:753–61.
Corcoran KA, Maren S. Hippocampal inactivation disrupts contextual retrieval of fear memory after extinction. J Neurosci. 2001;21:1720–6.
Corcoran KA, Maren S. Factors regulating the effects of hippocampal inactivation on renewal of conditional fear after extinction. Learn Mem. 2004;11:598–603.
Hobin JA, Ji J, Maren S. Ventral hippocampal muscimol disrupts context-specific fear memory retrieval after extinction in rats. Hippocampus. 2006;16:174–82.
Tayler KK, Tanaka KZ, Reijmers LG, Wiltgen BJ. Reactivation of neural ensembles during the retrieval of recent and remote memory. Curr Biol. 2013;23:99–106.
Denny CA, Kheirbek MA, Alba EL, Tanaka KF, Brachman RA, Laughman KB, et al. Hippocampal memory traces are differentially modulated by experience, time, and adult neurogenesis. Neuron. 2014;83:189–201.
Liu X, Ramirez S, Pang PT, Puryear CB, Govindarajan A, Deisseroth K, et al. Optogenetic stimulation of a hippocampal engram activates fear memory recall. Nature. 2012;484:381–5.
Ramirez S, Liu X, Lin PA, Suh J, Pignatelli M, Redondo RL, et al. Creating a false memory in the hippocampus. Science. 2013;341:387–91.
Martelotto LG. ‘Frankenstein’ protocol for nuclei isolation from fresh and frozen tissue for snRNAseq v2. 2019 May. Available from: https://www.protocols.io/view/frankenstein-protocol-for-nuclei-isolation-from-f-3fkgjkw.
Davis A, Gao R, Navin NE. SCOPIT: sample size calculations for single-cell sequencing experiments. BMC Bioinform. 2019;20:566.
Schmid KT, Höllbacher B, Cruceanu C, Böttcher A, Lickert H, Binder EB, et al. scPower accelerates and optimizes the design of multi-sample single cell transcriptomic studies. Nat Commun. 2021;12:6625.
Ewels P, Magnusson M, Lundin S, Käller M. MultiQC: summarize analysis results for multiple tools and samples in a single report. Bioinformatics. 2016;32:3047–8.
Wickham H, François R, Henry L, Müller K, Vaughan D. dplyr: a grammar of data manipulation. 2023. Available from: https://CRAN.R-project.org/package=dplyr.
Kassambara A. rstatix: pipe-friendly framework for basic statistical tests. 2023. Available from: https://CRAN.R-project.org/package=rstatix.
Lenth, Russel V. emmeans: estimated marginal means, aka least-squares means. 2023. Available from: https://CRAN.R-project.org/package=emmeans.
Olejnik S, Algina J. Generalized eta and omega squared statistics: measures of effect size for some common research designs. Psychol Methods. 2003;8:434–47.
Hao Y, Hao S, Andersen-Nissen E, Mauck WM, Zheng S, Butler A, et al. Integrated analysis of multimodal single-cell data. Cell. 2021;184:3573–.e29.
Ritchie ME, Phipson B, Wu D, Hu Y, Law CW, Shi W, et al. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47.
Zhou Y, Zhou B, Pache L, Chang M, Khodabakhshi AH, Tanaseichuk O, et al. Metascape provides a biologist-oriented resource for the analysis of systems-level datasets. Nat Commun. 2019;10:1523.
Cembrowski MS, Wang L, Sugino K, Shields BC, Spruston N. Hipposeq: a comprehensive RNA-seq database of gene expression in hippocampal principal neurons. eLife. 2016;5:e14997.
Olsen RHJ, Agam M, Davis MJ, Raber J. ApoE isoform-dependent deficits in extinction of contextual fear conditioning. Genes Brain Behav. 2012;11:806–12.
Johnson LA, Zuloaga DG, Bidiman E, Marzulla T, Weber S, Wahbeh H, et al. ApoE2 exaggerates PTSD-related behavioral, cognitive, and neuroendocrine alterations. Neuropsychopharmacology. 2015;40:2443–53.
Kovács KJ. Invited review c-Fos as a transcription factor: a stressful (re)view from a functional map. Neurochem Int. 1998;33:287–97.
Yap EL, Pettit NL, Davis CP, Nagy MA, Harmin DA, Golden E, et al. Bidirectional perisomatic inhibitory plasticity of a Fos neuronal network. Nature. 2021;590:115–21.
Moser MB, Moser EI. Functional differentiation in the hippocampus. Hippocampus. 1998;8:608–19.
Fanselow MS, Dong HW. Are the dorsal and ventral hippocampus functionally distinct structures? Neuron. 2010;1:7–19.
Kim JJ, Fanselow MS. Modality-specific retrograde amnesia of fear. Science. 1992;256:675–7.
Moser MB, Moser EI, Forrest E, Andersen P, Morris RG. Spatial learning with a minislab in the dorsal hippocampus. Proc Natl Acad Sci. 1995;92:9697–701.
Ferbinteanu J, McDonald RJ. Dorsal/ventral hippocampus, fornix, and conditioned place preference. Hippocampus. 2001;11:187–200.
Pothuizen HHJ, Zhang WN, Jongen-Relo AL, Feldon J, Yee BK. Dissociation of function between the dorsal and the ventral hippocampus in spatial learning abilities of the rat: a within-subject, within-task comparison of reference and working spatial memory. Eur J Neurosci. 2004;19:705–12.
Kjelstrup KG, Tuvnes FA, Steffenach HA, Murison R, Moser EI, Moser MB. Reduced fear expression after lesions of the ventral hippocampus. Proc Natl Acad Sci. 2002;99:10825–30.
Maren S, Holt WG. Hippocampus and pavlovian fear conditioning in rats: muscimol infusions into the ventral, but not dorsal, hippocampus impair the acquisition of conditional freezing to an auditory conditional stimulus. Behav Neurosci. 2004;118:97–110.
Pentkowski NS, Blanchard DC, Lever C, Litvin Y, Blanchard RJ. Effects of lesions to the dorsal and ventral hippocampus on defensive behaviors in rats. Eur J Neurosci. 2006;23:2185–96.
Esclassan F, Coutureau E, Di Scala G, Marchand AR. Differential contribution of dorsal and ventral hippocampus to trace and delay fear conditioning. Hippocampus. 2009;19:33–44.
Felix-Ortiz AC, Beyeler A, Seo C, Leppla CA, Wildes CP, Tye KM. BLA to vHPC inputs modulate anxiety-related behaviors. Neuron. 2013;79:658–64.
Swanson LW, Cowan WM. An autoradiographic study of the organization of the efferet connections of the hippocampal formation in the rat. J Comp Neurol. 1977;172:49–84.
Dong HW, Swanson LW, Chen L, Fanselow MS, Toga AW. Genomic–anatomic evidence for distinct functional domains in hippocampal field CA1. Proc Natl Acad Sci. 2009;106:11794–9.
Ciocchi S, Passecker J, Malagon-Vina H, Mikus N, Klausberger T. Selective information routing by ventral hippocampal CA1 projection neurons. Science. 2015;348:560–3.
Jimenez JC, Berry JE, Lim SC, Ong SK, Kheirbek MA, Hen R. Contextual fear memory retrieval by correlated ensembles of ventral CA1 neurons. Nat Commun. 2020;11:3492.
Jimenez JC, Su K, Goldberg AR, Luna VM, Biane JS, Ordek G, et al. Anxiety cells in a hippocampal-hypothalamic circuit. Neuron. 2018;97:670–.e6.
Parfitt GM, Nguyen R, Bang JY, Aqrabawi AJ, Tran MM, Seo DK, et al. Bidirectional control of anxiety-related behaviors in mice: role of inputs arising from the ventral hippocampus to the lateral septum and medial prefrontal cortex. Neuropsychopharmacology. 2017;42:1715–28.
Ramírez-Amaya V, Vazdarjanova A, Mikhael D, Rosi S, Worley PF, Barnes CA. Spatial exploration-induced arc mrna and protein expression: evidence for selective, network-specific reactivation. J Neurosci. 2005;25:1761–8.
Nasrouei S, Rattel JA, Liedlgruber M, Marksteiner J, Wilhelm FH. Fear acquisition and extinction deficits in amnestic mild cognitive impairment and early Alzheimer’s disease. Neurobiol Aging. 2020;87:26–34.
Hernandez CM, Jackson NL, Hernandez AR, McMahon LL. Impairments in fear extinction memory and basolateral amygdala plasticity in the TgF344-AD rat model of Alzheimer’s disease are distinct from nonpathological aging. eNeuro. 2022;9:ENEURO.0181-22.2022.
Bonardi C, de Pulford F, Jennings D, Pardon MC. A detailed analysis of the early context extinction deficits seen in APPswe/PS1dE9 female mice and their relevance to preclinical Alzheimer’s disease. Behav Brain Res. 2011;222:89–97.
Rattray I, Scullion GA, Soulby A, Kendall DA, Pardon MC. The occurrence of a deficit in contextual fear extinction in adult amyloid-over-expressing TASTPM mice is independent of the strength of conditioning but can be prevented by mild novel cage stress. Behav Brain Res. 2009;200:83–90.
Roheim PS, Carey M, Forte T, Vega GL. Apolipoproteins in human cerebrospinal fluid. Proc Natl Acad Sci. 1979;76:4646–9.
Vance JE, Hayashi H. Formation and function of apolipoprotein E-containing lipoproteins in the nervous system. Biochim Biophys Acta BBA - Mol Cell Biol Lipids. 2010;1801:806–18.
Pitas RE, Boyles JK, Lee SH, Hui D, Weisgraber KH. Lipoproteins and their receptors in the central nervous system. Characterization of the lipoproteins in cerebrospinal fluid and identification of apolipoprotein B,E(LDL) receptors in the brain. J Biol Chem. 1987;262:14352–60.
Freeman T, Roca V, Guggenheim F, Kimbrell T, Griffin WST. Neuropsychiatric associations of apolipoprotein E alleles in subjects with combat-related posttraumatic stress disorder. J Neuropsychiatry Clin Neurosci. 2005;17:541–3.
Kim TY, Chung HG, Shin HS, Kim SJ, Choi JH, Chung MY, et al. Apolipoprotein E gene polymorphism, alcohol use, and their interactions in combat-related posttraumatic stress disorder. Depress Anxiety. 2013;30:1194–201.
Weisgraber KH, Innerarity TL, Mahley RW. Abnormal lipoprotein receptor-binding activity of the human E apoprotein due to cysteine-arginine interchange at a single site. J Biol Chem. 1982;257:2518–21.
Wilson C, Mau T, Weisgraber KH, Wardell MR, Mahley RW, Agard DA. Salt bridge relay triggers defective LDL receptor binding by a mutant apolipoprotein. Structure. 1994;2:713–8.
Mulder M, Jansen PJ, Janssen BJA, van de Berg WDJ, van der Boom H, Havekes LM, et al. Low-density lipoprotein receptor-knockout mice display impaired spatial memory associated with a decreased synaptic density in the hippocampus. Neurobiol Dis. 2004;16:212–9.
de Oliveira J, Engel DF, de Paula GC, dos Santos DB, Lopes JB, Farina M, et al. High cholesterol diet exacerbates blood-brain barrier disruption in ldlr–/– mice: impact on cognitive function. J Alzheimers Dis. 2020;78:97–115.
de Oliveira J, Hort MA, Moreira ELG, Glaser V, Ribeiro-do-Valle RM, Prediger RD, et al. Positive correlation between elevated plasma cholesterol levels and cognitive impairments in LDL receptor knockout mice: relevance of cortico-cerebral mitochondrial dysfunction and oxidative stress. Neuroscience. 2011;197:99–106.
Mulder M, Koopmans G, Wassink G, Mansouri GA, Simard ML, Havekes LM, et al. LDL receptor deficiency results in decreased cell proliferation and presynaptic bouton density in the murine hippocampus. Neurosci Res. 2007;59:251–6.
Erwin SR, Sun W, Copeland M, Lindo S, Spruston N, Cembrowski MS. A sparse, spatially biased subtype of mature granule cell dominates recruitment in hippocampal-associated behaviors. Cell Rep. 2020;31:107551.
Cembrowski MS, Phillips MG, DiLisio SF, Shields BC, Winnubst J, Chandrashekar J, et al. Dissociable structural and functional hippocampal outputs via distinct subiculum cell classes. Cell. 2018;173:1280–.e18.
Gergues MM, Han KJ, Choi HS, Brown B, Clausing KJ, Turner VS, et al. Circuit and molecular architecture of a ventral hippocampal network. Nat Neurosci. 2020;23:1444–52.
Keinath AT, Mosser CA, Brandon MP. The representation of context in mouse hippocampus is preserved despite neural drift. Nat Commun. 2022;13:2415.
Delamare G, Zaki Y, Cai DJ, Clopath C. Drift of neural ensembles driven by slow fluctuations of intrinsic excitability. eLife. 2023;12:RP88053.
Acknowledgements
We thank Dr. Nihal Salem for guidance and the members of the Drew and Hofmann labs for discussion. Sequencing was performed by the Genomic Sequencing and Analysis Facility at UT Austin, Center for Biomedical Research Support (RRID# SCR_021713). Computational analyses were performed using the Biomedical Research Computing Facility at UT Austin, Center for Biomedical Research Support (RRID#: SCR_021979). This research was supported by a UT Austin Catalyst seed grant to HAH and MRD, U.S. National Institutes of Health grant R01 MH117426 to MRD, U.S. National Science Foundation grant IOS-1326187 to HAH, a U.S. Department of Justice graduate fellowship to IMC, and UT Austin Graduate School Summer Fellowships to JH and IMC, and a NIH T32 (MH106454) support to LAA.
Author information
Authors and Affiliations
Contributions
AZ: experiment design; data acquisition, analysis, interpretation, visualization; drafting and revising manuscript. JH: data analysis, interpretation, visualization; drafting and revising manuscript. IMC: experiment design; data acquisition; data analysis, interpretation, visualization; drafting and revising manuscript. LAA: data analysis, interpretation, visualization; drafting and revising manuscript. HAH: experiment design; funding acquisition; data interpretation; drafting and revising manuscript; project supervision MRD: experiment design; funding acquisition; data interpretation; drafting and revising manuscript; project supervision.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zuniga, A., Han, J., Miller-Crews, I. et al. Extinction training suppresses activity of fear memory ensembles across the hippocampus and alters transcriptomes of fear-encoding cells. Neuropsychopharmacol. (2024). https://doi.org/10.1038/s41386-024-01897-0
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1038/s41386-024-01897-0
- Springer Nature Switzerland AG