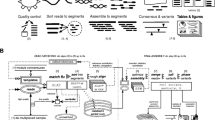
Abstract
Circular resequencing (CirSeq) is a novel technique for efficient and highly accurate next-generation sequencing (NGS) of RNA virus populations. The foundation of this approach is the circularization of fragmented viral RNAs, which are then redundantly encoded into tandem repeats by 'rolling-circle' reverse transcription. When sequenced, the redundant copies within each read are aligned to derive a consensus sequence of their initial RNA template. This process yields sequencing data with error rates far below the variant frequencies observed for RNA viruses, facilitating ultra-rare variant detection and accurate measurement of low-frequency variants. Although library preparation takes ∼5 d, the high-quality data generated by CirSeq simplifies downstream data analysis, making this approach substantially more tractable for experimentalists.




Similar content being viewed by others
References
Shendure, J. & Ji, H. Next-generation DNA sequencing. Nat. Biotechnol. 26, 1135–1145 (2008).
Dohm, J.C., Lottaz, C., Borodina, T. & Himmelbauer, H. Substantial biases in ultra-short-read data sets from high-throughput DNA sequencing. Nucleic Acids Res. 36, e105 (2008).
Hiatt, J.B., Patwardhan, R.P., Turner, E.H., Lee, C. & Shendure, J. Parallel, tag-directed assembly of locally derived short sequence reads. Nat. Methods 7, 119–122 (2010).
Schmitt, M.W. et al. Detection of ultra-rare mutations by next-generation sequencing. Proc. Natl. Acad. Sci. USA 109, 14508–14513 (2012).
Jabara, C.B., Jones, C.D., Roach, J., Anderson, J.A. & Swanstrom, R. Accurate sampling and deep sequencing of the HIV-1 protease gene using a Primer ID. Proc. Natl. Acad. Sci. USA 108, 20166–20171 (2011).
Kinde, I., Wu, J., Papadopoulos, N., Kinzler, K.W. & Vogelstein, B. Detection and quantification of rare mutations with massively parallel sequencing. Proc. Natl. Acad. Sci. USA 108, 9530–9535 (2011).
Acevedo, A., Brodsky, L. & Andino, R. Mutational and fitness landscapes of an RNA virus revealed through population sequencing. Nature 505, 686–690 (2014).
Lou, D.I. et al. High-throughput DNA sequencing errors are reduced by orders of magnitude using circle sequencing. Proc. Natl. Acad. Sci. USA 110, 19872–19877 (2013).
Langmead, B. & Salzberg, S.L. Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012).
Acknowledgements
We thank S. Taguwa and P. Ambrose for critical reading of the manuscript, and G. Schroth, M. Harrison, P. Wassam and T. Collins for technical advice. This work was financially supported by a National Science Foundation graduate research fellowship to A.A., and by National Institute of Allergy and Infectious Diseases (NIAID) grants AI091575, AI36178 and AI40085, as well as a Defense Advanced Research Projects Agency (DARPA) Prophecy grant to R.A.
Author information
Authors and Affiliations
Contributions
R.A. and A.A. conceived and designed the experiments; A.A. performed the experiments; and R.A. and A.A. wrote the manuscript.
Corresponding authors
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Rights and permissions
About this article
Cite this article
Acevedo, A., Andino, R. Library preparation for highly accurate population sequencing of RNA viruses. Nat Protoc 9, 1760–1769 (2014). https://doi.org/10.1038/nprot.2014.118
Published:
Issue Date:
DOI: https://doi.org/10.1038/nprot.2014.118
- Springer Nature Limited
This article is cited by
-
Evolutionary conservation of the fidelity of transcription
Nature Communications (2023)
-
The PPR domain of mitochondrial RNA polymerase is an exoribonuclease required for mtDNA replication in Drosophila melanogaster
Nature Cell Biology (2022)
-
An amplicon-based sequencing framework for accurately measuring intrahost virus diversity using PrimalSeq and iVar
Genome Biology (2019)
-
Hsp90 shapes protein and RNA evolution to balance trade-offs between protein stability and aggregation
Nature Communications (2018)
-
Poliovirus intrahost evolution is required to overcome tissue-specific innate immune responses
Nature Communications (2017)