Abstract
Plant oxylipins are derived from unsaturated fatty acids and play roles in plant growth and development as well as defence. Although recent studies have revealed that fatty acid metabolism is involved in systemic acquired resistance, the precise function of oxylipins in plant defence remains unknown. Here we report a cotton P450 gene SILENCE-INDUCED STEM NECROSIS (SSN), RNAi suppression of which causes a lesion mimic phenotype. SSN is also involved in jasmonate metabolism and the response to wounding. Fatty acid and oxylipin metabolite analysis showed that SSN overexpression causes hyperaccumulation of hydroxide and ketodiene fatty acids and reduced levels of 18:2 fatty acids, whereas silencing causes an imbalance in LOX (lipoxygenase) expression and excessive hydroperoxide fatty acid accumulation. We also show that an unknown oxylipin-derived factor is a putative mobile signal required for systemic cell death and hypothesize that SSN acts as a valve to regulate HR on pathogen infection.
Similar content being viewed by others
Introduction
Plants are constantly challenged by a barrage of microbes, but only a small proportion succeed in causing disease due to the well-established immune system of plants. Plant innate immune systems comprise complex signalling networks that generally include two modes, pathogen-associated molecular pattern-triggered immunity (PTI) and effector-triggered immunity (ETI)1. PTI is induced when pathogen-associated molecular patterns, which are conserved molecules such as flagellin, are perceived by extracellular receptors, the so-called pattern recognition receptors, such as FLS2 (ref. 2). However, when PTI is suppressed by pathogen effectors, and they are transported into the cell, plants can re-establish pathogen resistance using other defence modes, such as ETI3. ETI is induced when the avirulence effectors produced by a pathogen are recognized by the corresponding plant resistance proteins4. ETI is often accompanied by the hypersensitive response (HR), which includes an oxidative burst, cell wall lignification, phytoalexin accumulation and induction of cell death of infected cells and the cells that surround them, to prevent the pathogen from spreading5,6. HR is a form of programmed cell death (PCD) in plants7, which results in necrotic lesion formation, sealing the pathogen in a tomb of dead cells. This process is also associated with salicylic acid (SA) accumulation, which induces the expression of PATHOGENESIS-RELATED (PR) genes and eventually establishes systemic acquired resistance (SAR)8,9. Interestingly, SAR can be triggered in tobacco wild-type (WT) scion grafted onto an SA-deficient rootstock10. SAR has also been found to be blocked when SA methyl transferase was silenced in primary infected leaves, and SAR was induced in upper untreated leaves when lower leaves were treated with MeSA. Therefore, MeSA is a proposed SAR signal in tobacco11. Jasmonate (JA) rapidly accumulates in the phloem and exudates as the leaves are challenged with the avirulent Pseudomonas syringae; further, SAR can be mimicked by a foliar JA application and is abrogated in mutants with impaired JA synthesis or response12. In addition, the JA precursor 12-oxo-phytodienoic acid accumulates in SAR-induced potato plants13. Similarly, azelaic acid (AZA) is also derived from C18 fatty acids, and together with its induced protein AZELAIC ACID INDUCED 1 (AZI1) plays an important role in the systemic immune response14. Nevertheless, a recent study presents evidence that methyl salicylate and JA are non-essential for SAR in Arabidopsis, and AZA is a general marker for lipid peroxidation rather than a general immune signal15,16,17. Moreover, defective SAR in acyl carrier protein 4 (acp4) mutant plants was also not due to impaired salicylate or JA-mediated plant hormone signalling pathways but was associated with impaired leaf cuticlesFig. 4d). Further, recent research has shown that the JA pathway is important to V. dahliae infection51,52. Thus, we propose that JA, rather than SA, plays a key role in cotton V. dahliae resistance.
Methods
Plant materials and growth conditions
The cotton plants Gossypium barbadense cv. 7124, Gossypium hirsutum cv. Ji11 and Gossypium hirsutum cv. YZ1, as well as transgenic lines derived from YZ1 in these experiments, were cultivated in Wuhan, China under normal farming practices or grown in the greenhouse during the winter. The greenhouses were maintained at a temperature of 28–35/20–28 °C day/night. The roots, stems, cotyledons and leaves were collected from seedlings cultured in a growth chamber or in Hoagland’s solution. The collected materials were preserved at −80 °C until required for further analysis.
Plant treatments
For the JA/MeJA or AZA treatments using seedlings grown in Hoagland’s solution, 25 μM JA, 50 μM MeJA or 1 mM AZA (Sigma) was added to the Hoagland’s solution for the WT YZ1 plants (3 weeks), and the controls were added with ethanol to 0.01 or 0.2%. For the SA or ACC treatment, the cotton line YZ1 (3 weeks old) was grown in Hoagland’s solution, and 0.5 mM SA or 5 μM ACC (Sigma) was added; water was added to the controls. For the MeJA treatment in the medium, the WT and transgenic cotton lines were grown on a half-strength MS medium for 1.5 days and were then transferred to a mock treatment (0.01% ethanol) and media containing different concentrations of JA. For the AZA treatment on the medium, the WT and transgenic cotton lines were grown on a half-strength MS medium for 1.5 days; they were then transferred to a mock treatment (0.1% ethanol) and media containing the required AZA concentrations. For the glycerol treatment on the medium, the WT and transgenic cotton lines were grown on half-strength MS medium for 1.5 days; they were then transferred to a half-strength MS medium supplemented with or without 0.5% glycerol. For the FA treatment, FA stock solutions (200 mM) were prepared in 80% ethanol. Working solutions (10 mM) were prepared in deionized water with the surfactant SilwetL77 (0.02%). Mock solutions were prepared with ethanol (1%) and surfactant (0.02%) in deionized water. Cotyledons from the WT cotton line YZ1 (1 week old) were grown in a conical flask and sprayed with mock treatment or 10 mM various FAs (OA, LA and ALA), respectively. The tissues were then harvested at the indicated time intervals for RNA extraction.
Wound treatment
For silenced plants, the taproots of 3-week-old plants were grown in Hoagland’s solution and cut to lengths of ~1 cm using scissors. At various time points after wounding, the roots (four or five plants) were harvested, immediately frozen in liquid nitrogen and stored at −80 °C until they were used for RNA or JA extraction. For the overexpressing plants, fully expanded true leaves of 4-week-old plants were wounded to ~40% of the leaf area by crushing the leaf across the midrib using a haemostat. The leaves (four or five plants) were harvested as described above.
Pathogen infection
G. barbadense cv. 7124 and G. hirsutum cv. Ji11 seeds were grown in commercial sterilized soil for 3 weeks. The plants were inoculated with the V. dahliae strain V991, as described previously24. The plant roots were harvested for RNA extraction at 0, 1, 4, 12 and 24 h after inoculation. The control plants were treated with water and sampled at the same time points. Roots from five individual seedlings were collected for each treatment at each sampling time point. Roots from 3-week-old Hoagland’s solution-cultured WT and transgenic plant seedlings were separately wounded with scissors and dip-inoculated with the V. dahliae strain V991 to observe the disease symptoms.
SSN cloning and sequence analysis
Total RNA was extracted from cv. YZ1 or cv. 7124 roots using the guanidine thiocyanate method53. The first strand of cDNA was synthesized using the SuperScript III reverse transcriptase (Invitrogen, Carlsbad, CA, USA). The expressed sequence tag sequence was isolated from a suppression subtractive hybridization library of cotton line 7124 (ref. 24). The full-length sequence was obtained through 5′- and 3′-rapid amplification of the cDNA end (5′- and 3′-RACE) in accordance with the GeneRacer Kit user manual (Invitrogen) using cv. YZ1 cDNA as the PCR template. The ORF was predicted using ORF Finder ( http://www.ncbi.nlm.nih.gov/gorf/gorf.html). Three genes with highly similar sequences were found during the sequencing process, which are referred to as GhCYP82D1, GhCYP82D2 and GhCYP82D3. The primer sequences are listed in Supplementary Table 2. A sequence similarity analysis was performed using BOXSHADE software ( http://www.ch.embnet.org/software/BOX_form.html), and the sequence alignments were completed using the ClustalX and MEGA5 software with the neighbour-joining method. The GhCYP82D1 and GhCYP82D2 promoter sequences were obtained through DNA walking in accordance with the Genome Walker Universal Kit User Manual (Clontech) using cv. YZ1 DNA as the PCR template.
Plasmid construction and plant transformation
The conserved region of the three SSN genes and 3′-UTR-specific regions of each gene were individually selected as the RNAi target and cloned into the RNAi vector pHellsgate 4 using the gateway system. Full-length SSN1 and SSN2 genes were amplified from the cv. YZ1 cDNA and cloned into pK2GW7,0 (Ghent University). The SSN1 and SSN2 promoters were fused with the GUS reporter gene in pGWB433 (Research Institute of Molecular Genetics, Shimane University, Matsue, Japan). The expression constructs were introduced into cv. YZ1 through the Agrobacterium tumefaciens strains LBA4404 or EHA105 (ref. 54). The TRV vectors were constructed, and the Agrobacterium tumefaciens (GV3101) were prepared for virus-induced gene silencing in accordance with a previous study51. The sequences used to construct TRV:GhLOX1 and TRV:GhLOX2 were amplified from the cv. YZ1 cDNA. The PCR fragments were digested with BamHI and KpnI and then ligated into the TRV:00 plasmid. The primer sequences are listed in Supplementary Table 2. The vectors were used to transform A. tumefaciens through electroporation. The A. tumefaciens with TRV vectors were infiltrated into the 10-day-old YZ1 seedling cotyledons. The seedlings were then grown at 25 °C with a 16/8 h light/dark photoperiod in an incubator.
Illumina sequencing analysis
Total RNA was extracted using a plant total RNA kit (Sigma, St Louis, MO, USA) from 6-day-old Ri15 line and WT control seedling roots grown under identical conditions on a half-strength MS medium. RNA sequencing and data analysis were performed by the Bei**g Genomics Institute (BGI, Shenzhen, China) using the Illumina Genome Analyzer. Briefly, the cDNA was digested with NlaIII and then ligated with the first adaptor containing the recognition site of MmeI, a type II endonuclease that cleaves at sites 21 bp from the recognition site. After digestion by MmeI, the transcripts were ligated with the second adaptor. Single-end sequencing mode was used and the read length was 49 bp. A sequence data set was constructed from cotton unigenes using NCBI ( http://www.ncbi.nlm.nih.gov/unigene/?term=txid3633[Organism:exp]), which was used as the reference database. SOAPaligner/soap2 method was used to map reads and then the expression level of each gene was normalized to RPKM (reads per kb per million reads. The differential gene expression between WT and Ri15 was determined by taking the log2 ratio of RPKM. The Audic and Claverie method was used for normalization procedures and statistical data analysis55. FDR was used to determine the threshold of P value in multiple test and analysis. The RPKM>15 in any one sample and the absolute value of log2 ratio >1 based on the FDR<0.001 were used as the threshold to judge the significance of gene expression difference.
Histochemical assay
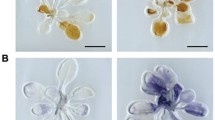
For GUS detection, fresh tissues were collected from the SSN1 promoter::GUS plants and SSN2 promoter::GUS plants, dipped into pre-chilled 80% (v/v) acetone for 30 min, infiltrated into a staining solution under a vacuum for 15 min and then moved to 37 °C for 6 h. The staining solution was composed of 0.9 g l−1 5-bromo-4-chloro-3-indolyl-b-glucuronic acid, 50 mM sodium phosphate buffer (pH 7.0), 20% (v/v) methanol and 100 mg l−1 chloromycetin. The samples were successively washed with 75% ethanol and then examined and photographed with a Nikon D40 camera (Japan). For H2O2 detection, the stems without a lesion phenotype in SSN-RNAi and WT plants were sampled for H2O2 measurements as described previously56. For DAB staining, SSN-RNAi plant cotyledons were incubated in 1 mg ml−1 pH 3.8 DAB-HCl (Sigma-Aldrich, USA) in the dark for 8 h. The cotyledons were then cleared by boiling in alcoholic lactophenol (95% ethanol:lactophenol, 2:1 v/v) for 20 min. The reddish colour of the cotyledons was used as evidence of H2O2 and visualized using a Nikon D40 camera (Japan). For 2′,7′-DCFDA staining, the hypocotyls of each lines were excised from 9-day-old seedlings, which have no lesion phenotype on stems on media and cut into about 5-mm segments, then incubated for 30 min in the dark at 30 °C in 2′,7′-DCFDA diluted with phosphate buffer solution to 10 μM. The images were observed and recorded using an Olympus light microscope equipped with an Olympus U-PMTVC adapter and a Leica DC300F camera. For section observation, the hypocotyls of each lines were excised from 9-day-old seedlings, which have the weak lesion spot on stems on media and cut into 5–7 mm segments, and then were fixed in FAA solution (formaldehyde:acetic acid: 70% alcohol:water, 1:1:10:8) overnight at 4 °C. Samples were dehydrated in a progressive series of ethanol dilution, infiltrated in chloroform overnight at 37 °C and embedded in paraffin wax (melting point 56 °C). Sections (8 μm) were prepared using a rotary microtome (KD 2058, KEDEE, China) and stained with Safranin fast green. The sections were observed and photographed under a photomicroscope (DM2500, Leica, Wetzlar, Germany). More than 10 samples of each line were analysed above.
Transmission electron microscopy scanning
WT and transgenic seedling cotyledons were grown for 6 days under identical conditions, cut into 1 mm2 pieces with a scalpel, prefixed in the fixing solution (2.5% glutaraldehyde adjusted to pH 7.4 with 0.1 M phosphate buffer and fixed in 2% OsO4 in the same buffer) and then cut into smaller pieces with scissors. Ultrathin sections were obtained using a Leica UC6 ultramicrotome (Leica, Germany) and stained with uranyl acetate then with lead citrate. The images were observed and recorded using a HITACHI H-7650 transmission electron microscope (Hitachi High-tech, Ibaraki-ken, Honshu, Japan) at 80 KV and a Gatan 832 CCD camera (Gatan, Pleasanton, CA).
Quantitative real-time reverse transcription-PCR
The total RNA was isolated as previously described57. Tissues were ground in a mortar with liquid nitrogen and ice-cold extraction buffer containing 1% β-mercaptoethanol was added and mixed completely by inverting the tube. The supernatant was purified by phenol and chloroform. The RNA was precipitated by isopropanol and sodium acetate (3 M) and then washed by 75% ethanol. Air-dried RNA was dissolved in diethylpyrocarbonate-treated water. The RNA was reverse transcribed to cDNA using the SuperScript III reverse transcriptase (Invitrogen). Quantitative real-time (qRT) PCR was performed using the ABI Prism 7000 system (Applied Biosystems, Foster City, CA, USA). For qRT-PCR analysis, at least 5–10 plants of every line or treatment were sampled for each independent biological replicate. Gene sequences were obtained from the public NCBI UniGene data bank. The primers are listed in Supplementary Table 2. Four technical replicates or three biological replicates for each experiment were performed. Error bars represent the s.d. UBQ7 was amplified as a control. The expression value of all genes was normalized by referring to UBQ7 as ‘1’.
Southern, northern and western blotting
Genomic DNA was extracted from young leaves of transgenic cotton lines using the plant genomic DNA kit DP305 (Tiangen Biotech, Bei**g). The NPTII gene was used as a probe for Southern blotting to detect transgene insertion. Total RNA from the leaves of 3-week-old seedlings was extracted as described above; a PR1 fragment was used as the probe for northern blot analysis in SSN-RNAi plants. Procedures for hybridization and washing the membrane were as previously reported56. The primer sequences are listed in Supplementary Table 2. For western blotting, a specific amino acid sequence from the same region of SSN1 and SSN2 (residues 272–286 DHRKGGRWDENKKEK) was used as the antigen through commercial synthesis; the antibody was prepared by Neweast Bioscience (Wuhan, China). Anti-histone 3 (ab1791, Rabbit polyclonal to Histone H3—ChIP Grade, ABcam, San Francisco, CA) was used as an endogenous standard. Total protein was extracted from WT, SSN-RNAi and overexpressing seedlings using the extraction buffer. Protein concentrations were determined using the Bradford assay. Western blot experiments were performed as described previously58. The primary antibody was diluted at 1:5,000 for SSN probing or 1:10,000 for histone probing. A secondary anti-rabbit horseradish peroxidase labelled IgG was subsequently added. Signal of protein was detected using the Pierce Supersignal West Pico kit (Thermo Scientific, Rockford, IL).
Transient expression and subcellular localization
Full-length SSN1 cDNA without a stop codon was isolated using the primers forward, 5′-GGGGACAAGTTTGTACAAAAAAGCAGGCTTCATGGATCTTCTTGATTTCTCCACT-3′, and reverse, 5′-GGGGACCACTTTGTACAAGAAAGCTGGGTTGTTATAGAGCTCAGGAGCAAGGC-3′, carrying the attB adapter sites and recombined into the vector pMDC83 (ref. 59) by BP and LR recombination reactions (Gateway Technology); the resulting construct consisted of SSN1 fused to the green fluorescent protein N terminus controlled by the CaMV 35S promoter. An RFP fusion with the chaperone-binding protein BiP was used as an endoplasmic reticulum marker protein. The fusion constructs were introduced into Nicotiana benthamiana protoplasts isolated from seedling leaves by polyethylene glycol/calcium-mediated transformation60. Fluorescence microscopy was performed using a Leica TCS SP2 confocal spectral microscope (Leica, Heidelberg, Germany).
Lipid analysis
WT and transgenic plant cotyledons were quickly immersed in 3.0 ml 75 °C isopropanol with 0.01% butylated hydroxytoluene for 15 min. The mixture was vortexed after adding 1.5 ml chloroform and 0.6 ml water; it was then agitated (on shaking incubator) at room temperature for 1 h. The lipid extracts were transferred to glass screw-cap (Teflon-lined) tubes. Next, 4.0 ml chloroform/methanol (2:1) with 0.01% butylated hydroxytoluene was added before shaking for 30 min. The above procedure was repeated for all samples until the leaves of each sample became white. The process typically required approximately five extractions, including the isopropanol extraction. KCl (1 M, 1.0 ml) was added to the combined extract, the sample mixed by vortexing or shaking, then centrifuged to break up the phases and the upper phase was discarded. Water (2.0 ml) was added, vortexed or shaken, centrifuged and the upper phase was discarded. Extracted leaves were dried at 80 °C in an oven overnight for dry weight determination to four decimal places (0.0000, g). Samples were analysed by the KLRC (Kansas Lipidomics Research Center). The free fatty acids were analyzed in the (Applied Biosystems) API4000 triple quad mass spec using the negative MS1 scan mode. The source temperature was 100 °C, the desolvation temperature was 250 °C, 2.8 kV was applied to the electrospray capillary, the cone energy was 40 V and argon was used as the collision gas at 1.7 e−3 mBar as measured on the gauge in the collision cell line.
Phytohormone and oxylipin quantitation
For JA analysis, the samples (100–200 mg) were extracted twice with 80% cold methanol (v/v) overnight at 4 °C. To each sample was added 10 ng (±)-9,10-dihydro-JA (Olchemim) as an internal standard. The combined extract was evaporated to the aqueous phase with N2, dissolved in 0.4 ml methanol and then filtered using a syringe-facilitated filter (Nylon 66; ** Teng Experiment Equipment Co., Tian**, China). The samples were stored at −80 °C before the measurements. The JA levels were quantified using an HPLC-MS/MS system (AB SCIEX Triple Quad 5500 LC/MS/MS) with JA (Sigma) as the external standards. For JA-Ile and SA analysis, the JA-Ile and SA levels were quantified with JA-Ile and SA (Sigma) as the external standards. To estimate the oxylipins levels, the samples were obtained and extracted as described for phytohormone analysis. The supernatants were analysed using an HPLC-MS/MS system (AB SCIEX Triple Quad 5500 LC/MS/MS system) with 9- or 13-HPOD, 9- or 13-HPOT, 9- or 13-HOD, 9- or 13-HOT or 9- or 13-KOD (Cayman Chemical Co) as the external standard.
Additional information
Accession codes: cDNA sequences have been deposited in the NCBI GenBank database with the following accession codes: GhCYP82D1 (SSN1), KJ704109; cDNA of GhCYP82D2 (SSN2), KJ704110; cDNA of GhCYP82D3 (SSN3), KJ704111. Raw RNA-seq data have been deposited in the Sequence Read Archive (SRA) under accession codes SRR1569164.
How to cite this article: Sun, L. et al. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat. Commun. 5:5372 doi: 10.1038/ncomms6372 (2014).
References
Thomma, B. P., Nürnberger, T. & Joosten, M. H. Of PAMPs and effectors: the blurred PTI-ETI dichotomy. Plant Cell 23, 4–15 (2011).
Lu, D. et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc. Natl Acad. Sci. USA 107, 496–501 (2010).
Nomura, K. et al. Effector-triggered immunity blocks pathogen degradation of an immunity-associated vesicle traffic regulator in Arabidopsis. Proc. Natl Acad. Sci. USA 108, 10774–10779 (2011).
Bonas, U. & Lahaye, T. Plant disease resistance triggered by pathogen-derived molecules: refined models of specific recognition. Curr. Opin. Microbiol. 5, 44–50 (2002).
Hammond-Kosack, K. E. & Jones, J. D. Resistance gene-dependent plant defense responses. Plant Cell 8, 1773 (1996).
Tsuji, J., Jackson, E. P., Gage, D. A., Hammerschmidt, R. & Somerville, S. C. Phytoalexin accumulation in Arabidopsis thaliana during the hypersensitive reaction to Pseudomonas syringae pv. syringae. Plant Physiol. 98, 1304–1309 (1992).
Liu, Y. et al. Autophagy regulates programmed cell death during the plant innate immune response. Cell 121, 567–577 (2005).
Métraux, J. et al. Increase in salicylic acid at the onset of systemic acquired resistance in cucumber. Science 250, 1004–1006 (1990).
Durrant, W. & Dong, X. Systemic acquired resistance. Annu. Rev. Phytopathol. 42, 185–209 (2004).
Pallas, J. A., Paiva, N. L., Lamb, C. & Dixon, R. A. Tobacco plants epigenetically suppressed in phenylalanine ammonia‐lyase expression do not develop systemic acquired resistance in response to infection by tobacco mosaic virus. Plant J. 10, 281–293 (1996).
Park, S. W., Kaimoyo, E., Kumar, D., Mosher, S. & Klessig, D. F. Methyl salicylate is a critical mobile signal for plant systemic acquired resistance. Science 318, 113–116 (2007).
Truman, W., Bennett, M. H., Kubigsteltig, I., Turnbull, C. & Grant, M. Arabidopsis systemic immunity uses conserved defense signaling pathways and is mediated by jasmonates. Proc. Natl Acad. Sci. USA 104, 1075–1080 (2007).
Landgraf, P., Feussner, I., Hunger, A., Scheel, D. & Rosahl, S. Systemic accumulation of 12-oxo-phytodienoic acid in SAR-induced potato plants. Eur. J. Plant Pathol. 108, 279–283 (2002).
Jung, H. W., Tschaplinski, T. J., Wang, L., Glazebrook, J. & Greenberg, J. T. Priming in systemic plant immunity. Science 324, 89–91 (2009).
Attaran, E., Zeier, T. E., Griebel, T. & Zeier, J. Methyl salicylate production and jasmonate signaling are not essential for systemic acquired resistance in Arabidopsis. Plant Cell 21, 954–971 (2009).
Zoeller, M. et al. Lipid profiling of the Arabidopsis hypersensitive response reveals specific lipid peroxidation and fragmentation processes: biogenesis of pimelic and azelaic acid. Plant Physiol. 160, 365–378 (2012).
Chaturvedi, R. et al. Plastid ω3‐fatty acid desaturase‐dependent accumulation of a systemic acquired resistance inducing activity in petiole exudates of Arabidopsis thaliana is independent of jasmonic acid. Plant J. 54, 106–117 (2008).
**a, Y. et al. An intact cuticle in distal tissues is essential for the induction of systemic acquired resistance in plants. Cell Host Microbe 5, 151–165 (2009).
Maldonado, A. M., Doerner, P., Dixon, R. A., Lamb, C. J. & Cameron, R. K. A putative lipid transfer protein involved in systemic resistance signalling in Arabidopsis. Nature 419, 399–403 (2002).
Chanda, B. et al. Glycerol-3-phosphate is a critical mobile inducer of systemic immunity in plants. Nat. Genet. 43, 421–427 (2011).
Chaturvedi, R. et al. An abietane diterpenoid is a potent activator of systemic acquired resistance. Plant J. 71, 161–172 (2012).
Návarová, H., Bernsdorff, F., Döring, A. C. & Zeier, J. Pipecolic acid, an endogenous mediator of defense amplification and priming, is a critical regulator of inducible plant immunity. Plant Cell 24, 5123–5141 (2012).
Nandi, A., Welti, R. & Shah, J. The Arabidopsis thaliana dihydroxyacetone phosphate reductase gene SUPPRESSOR OF FATTY ACID DESATURASE DEFICIENCY1 is required for glycerolipid metabolism and for the activation of systemic acquired resistance. Plant Cell 16, 465–477 (2004).
Xu, L. et al. Differential gene expression in cotton defence response to Verticillium dahliae by SSH. J. Phytopathol. 159, 606–615 (2011).
Apel, K. & Hirt, H. Reactive oxygen species: metabolism, oxidative stress, and signal transduction. Annu. Rev. Plant Biol. 55, 373–399 (2004).
Sels, J., Mathys, J., De Coninck, B., Cammue, B. & De Bolle, M. F. Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950 (2008).
Alvarez, M. E. Salicylic acid in the machinery of hypersensitive cell death and disease resistance. Plant Mol. Biol. 44, 429–442 (2000).
Bell, E., Creelman, R. A. & Mullet, J. E. A chloroplast lipoxygenase is required for wound-induced jasmonic acid accumulation in Arabidopsis. Proc. Natl Acad. Sci. USA 92, 8675–8679 (1995).
Bertoni, G. Oxylipins and plant palatability. Plant Cell 24, 1305–1305 (2012).
Montillet, J. L. et al. An abscisic acid-independent oxylipin pathway controls stomatal closure and immune defense in Arabidopsis. PLoS Biol. 11, e1001513 (2013).
Jalloul, A. et al. Lipid peroxidation in cotton: Xanthomonas interactions and the role of lipoxygenases during the hypersensitive reaction. Plant J. 32, 1–12 (2002).
Ellis, C. & Turner, J. G. The Arabidopsis mutant cev1 has constitutively active jasmonate and ethylene signal pathways and enhanced resistance to pathogens. Plant Cell 13, 1025–1033 (2001).
Montillet, J. L. et al. Fatty acid hydroperoxides and H2O2 in the execution of hypersensitive cell death in tobacco leaves. Plant Physiol. 138, 1516–1526 (2005).
Fan, J., Yan, C. & Xu, C. Phospholipid: diacylglycerol acyltransferase‐mediated triacylglycerol biosynthesis is crucial for protection against fatty acid‐induced cell death in growing tissues of Arabidopsis. Plant J. 76, 930–942 (2013).
Marmey, P. et al. The 9-lipoxygenase GhLOX1 gene is associated with the hypersensitive reaction of cotton Gossypium hirsutum to Xanthomonas campestris pv. malvacearum. Plant Physiol Biochem. 45, 596–606 (2007).
Hwang, I. S. & Hwang, B. K. The pepper 9-lipoxygenase gene CaLOX1 functions in defense and cell death responses to microbial pathogens. Plant Physiol. 152, 948–967 (2010).
García-Marcos, A., Pacheco, R., Manzano, A., Aguilar, E. & Tenllado, F. Oxylipin biosynthesis genes positively regulate programmed cell death during compatible infections with the synergistic pair Potato virus X-Potato virus Y and Tomato Spotted Wilt Virus. J. Virol. 87, 5769–5783 (2013).
Nelson, D. R., Schuler, M. A., Paquette, S. M., Werck-Reichhart, D. & Bak, S. Comparative genomics of rice and Arabidopsis. Analysis of 727 cytochrome P450 genes and pseudogenes from a monocot and a dicot. Plant Physiol. 135, 756–772 (2004).
Frank, M. R., Deyneka, J. M. & Schuler, M. A. Cloning of wound-induced cytochrome P450 monooxygenases expressed in pea. Plant Physiol. 110, 1035–1046 (1996).
Schopfer, C. & Ebel, J. Identification of elicitor-induced cytochrome P450s of soybean (Glycine max L.) using differential display of mRNA. Mol. Gen. Genet. 258, 315–322 (1998).
Ralston, L. et al. Cloning, Heterologous expression, and functional characterization of 5- epi-aristolochene-1, 3-dihydroxylase from tobacco (Nicotiana tabacum). Arch. Biochem. Biophys. 393, 222–235 (2001).
Liu, F. et al. The Arabidopsis P450 protein CYP82C2 modulates jasmonate-induced root growth inhibition, defense gene expression and indole glucosinolate biosynthesis. Cell Res. 20, 539–552 (2010).
Lee, S. et al. Herbivore-induced and floral homoterpene volatiles are biosynthesized by a single P450 enzyme (CYP82G1) in Arabidopsis. Proc. Natl Acad. Sci. USA 107, 21205–21210 (2010).
Collazo, C., Chacón, O. & Borrás, O. Programmed cell death in plants resembles apoptosis of animals. Biotecnol. Apl. 23, 1–10 (2006).
Maccarrone, M., Melino, G. & Finazzi-Agro, A. Lipoxygenases and their involvement in programmed cell death. Cell Death Differ. 8, 776–784 (2001).
Higgins, L. J. & Rutledge, J. C. Inflammation associated with the postprandial lipolysis of triglyceriderich lipoproteins by lipoprotein lipase. Curr. Atheroscler. Rep. 11, 199–205 (2009).
Rustérucci, C. et al. Involvement of lipoxygenase-dependent production of fatty acid hydroperoxides in the development of the hypersensitive cell death induced by cryptogein on tobacco leaves. J. Biol. Chem. 274, 36446–36455 (1999).
Knight, V. I. et al. Hydroperoxides of fatty acids induce programmed cell death in tomato protoplasts. Physiol. Mol. Plant Pathol. 59, 277–286 (2001).
Göbel, C. & Feussner, I. Methods for the analysis of oxylipins in plants. Phytochemistry 70, 1485–1503 (2009).
Klosterman, S. J. et al. Comparative genomics yields insights into niche adaptation of plant vascular wilt pathogens. PLoS Pathog. 7, e1002137 (2011).
Gao, W. et al. Proteomic and virus-induced gene silencing (VIGS) analyses reveal that gossypol, brassinosteroids, and jasmonic acid contribute to the resistance of cotton to Verticillium dahliae. Mol. Cell. Proteomics 12, 3690–3703 (2013).
Fradin, E. F. et al. Interfamily transfer of tomato Ve1 mediates Verticillium resistance in Arabidopsis. Plant Physiol. 156, 2255–2265 (2011).
Zhu, L. F., Tu, L. L., Zeng, F. C., Liu, D. Q. & Zhang, X. L. An improved simple protocol for isolation of high quality RNA from Gossypium spp. suitable for cDNA library construction. Acta Agron. Sinica 31, 1657–1659 (2005).
**, S. X. et al. Identification of a novel elite genotype for in vitro culture and genetic transformation of cotton. Biol. Plantarum 50, 519–524 (2006).
Audic, S. & Claverie, J. M. The significance of digital gene expression profiles. Genome Res. 7, 986–995 (1997).
Deng, F. et al. GbPDF1 is involved in cotton fiber initiation via the core cis-element HDZIP2ATATHB2. Plant Physiol. 158, 890–904 (2012).
Liu, D., Zhang, X., Tu, L., Zhu, L. & Guo, X. Isolation by suppression-subtractive hybridization of genes preferentially expressed during early and late fiber development stages in cotton. Mol. Biol. 40, 741–749 (2006).
Hu, L., Yang, X., Yuan, D., Zeng, F. & Zhang, X. GhHmgB3 deficiency deregulates proliferation and differentiation of cells during somatic embryogenesis in cotton. Plant Biotechnol. J. 9, 1038–1048 (2011).
Curtis, M. D. & Grossniklaus, U. A gateway cloning vector set for high-throughput functional analysis of genes in planta. Plant Physiol. 133, 462–469 (2003).
Yoo, S. D., Cho, Y. H. & Sheen, J. Arabidopsis mesophyll protoplasts: a versatile cell system for transient gene expression analysis. Nat. Protoc. 2, 1565–1572 (2007).
Acknowledgements
We are grateful to KLRC (Kansas Lipidomics Research Center) for lipid quantification, D. Li and H. Liu (National Key Laboratory of Crop Genetic Improvement, China) for hormone and oxylipin quantification, Professor Q. Zhang (National Key Laboratory of Crop Genetic Improvement, Huazhong Agricultural University, Wuhan, China) for providing the endoplasmic reticulum marker, J. Cao and Y. Tan (Huazhong Agricultural University, China) for assistance with transmission electron microscopy, Professor J. Yao for hel** with section observation and Professor K. Lindsey (School of Biological and Biomedical Sciences, University of Durham) for revising the manuscript. This research was supported by the National Natural Science Foundation of China (31271772) and National High-Tech Program (2013AA102601-4).
Author information
Authors and Affiliations
Contributions
X.Z. and L.Z. conceived and designed the experiments; L.X. constructed the suppression subtractive hybridization cDNA library; D.Y. annotated the RNA-Seq data; L.S. performed the experiments, analysed the data and wrote the paper and X.Z. and L.M. corrected the manuscript. All of the authors discussed the results and commented on the manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare no competing financial interests.
Supplementary information
Supplementary Information
Supplementary Figures 1-16, Supplementary Tables 1-2. (PDF 4797 kb)
Rights and permissions
This work is licensed under a Creative Commons Attribution 4.0 International License. The images or other third party material in this article are included in the article’s Creative Commons license, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons license, users will need to obtain permission from the license holder to reproduce the material. To view a copy of this license, visit http://creativecommons.org/licenses/by/4.0/
About this article
Cite this article
Sun, L., Zhu, L., Xu, L. et al. Cotton cytochrome P450 CYP82D regulates systemic cell death by modulating the octadecanoid pathway. Nat Commun 5, 5372 (2014). https://doi.org/10.1038/ncomms6372
Received:
Accepted:
Published:
DOI: https://doi.org/10.1038/ncomms6372
- Springer Nature Limited
This article is cited by
-
Overexpression of cucumber CYP82D47 enhances resistance to powdery mildew and Fusarium oxysporum f. sp. cucumerinum
Functional & Integrative Genomics (2024)
-
Polyethyleneimine-coated MXene quantum dots improve cotton tolerance to Verticillium dahliae by maintaining ROS homeostasis
Nature Communications (2023)
-
Rice (Oryza sativa L.) cytochrome P450 protein 716A subfamily CYP716A16 regulates disease resistance
BMC Genomics (2022)
-
Advances and prospects of genetic map** of Verticillium wilt resistance in cotton
Journal of Cotton Research (2022)
-
ATP-citrate lyase B (ACLB) negatively affects cell death and resistance to Verticillium wilt
BMC Plant Biology (2022)