Abstract
The transcription factor nuclear factor-kappaB (NF-κB) is constitutively active in several cancers and is a target of therapeutic development. We recently developed dimethylaminoparthenolide (DMAPT), a clinical grade water-soluble analog of parthenolide, as a potent inhibitor of NF-κB and demonstrated in vitro and in vivo anti-tumor activities in multiple cancers. In this study, we show DMAPT is an epigenetic modulator functioning in an NF-κB-dependent and -independent manner. DMAPT-mediated NF-κB inhibition resulted in elevated histone H3K36 trimethylation (H3K36me3), which could be recapitulated through genetic ablation of the p65 subunit of NF-κB or inhibitor-of-kappaB alpha super-repressor overexpression. DMAPT treatment and p65 ablation increased the levels of H3K36 trimethylases NSD1 (KMT3B) and SETD2 (KMT3A), suggesting that NF-κB directly represses their expression and that lower H3K36me3 is an epigenetic marker of constitutive NF-κB activity. Overexpression of a constitutively active p65 subunit of NF-κB reduced NSD1 and H3K36me3 levels. NSD1 is essential for DMAPT-induced expression of pro-apoptotic BIM, indicating a functional link between epigenetic modification and gene expression. Interestingly, we observed enhanced H4K20 trimethylation and induction of H4K20 trimethylase KMT5C in DMAPT-treated cells independent of NF-κB inhibition. These results add KMT5C to the list NF-κB-independent epigenetic targets of parthenolide, which include previously described histone deacetylase 1 (HDAC-1) and DNA methyltransferase 1. As NSD1 and SETD2 are known tumor suppressors and loss of H4K20 trimethylation is an early event in cancer progression, which contributes to genomic instability, we propose DMAPT as a potent pharmacologic agent that can reverse NF-κB-dependent and -independent cancer-specific epigenetic abnormalities.
Similar content being viewed by others
Main
Epigenetics is defined as heritable changes in gene expression mediated mostly by DNA methylation and histone tail modifications without changes in DNA sequence.1 Epigenetic abnormalities in cancer lead to reprogramming of gene expression resembling embryonic stem cells, loss of tumor suppressors, or reactivation of oncofetal genes.2, 3 For example, enhanced histone 3 lysine 27 trimethylation (H3K27me3), mediated by a complex of proteins, including the histone methyltransferase EZH2, causes the silencing of tumor-suppressor genes.4, 5
Similar to H3K27me3, histone 3 lysine 36 trimethylation (H3K36me3) has a diverse role in chromatin structure and function.6 H3K36me3 regulates transcription of active euchromatin, alternative splicing, DNA repair, and transmission of the memory of gene expression from parents to offspring.7 At least eight enzymes methylate H3K36. Nuclear receptor-binding SET domain protein 1 (NSD1; lysine methyltransferases 3B (KMT3B)) and NSD2 are involved in monomethylation and dimethylation, whereas SET domain containing 2 (SETD2; KMT3A) trimethylates H3K36.6 However, depletion of NSD1 in certain cells leads to loss of H3K36me3 and altered gene expression.8, 9
NSD1 functions as a tumor suppressor in several cancers. For example, epigenetic silencing of NSD1 is observed in neuroblastoma.9 NSD1 mutation/deletion is observed in bladder cancer.10 NSD1 is one of the genes essential for anti-estrogen sensitivity in breast cancer.11 NSD1 is also described as an oncogene in acute myeloid leukemia in which a chromosomal translocation creates nucleoporin 98 kDa (NUP98):NSD1 fusion protein, and the expression is driven by NUP98 regulatory elements.12 NSD2 is an oncogene, whereas SETD2 is inactivated through mutation/deletion in multiple cancers.10, 13, 14 Interestingly, H3K36me3 and H3K27me3 on a gene may be mutually exclusive, suggesting distinct role of modulators of these modifications on gene expression.15
Loss of histone 4 lysine 20 trimethylation (H4K20me3) is a hallmark of cancer.16 The histone methyltransferases KMT5B (SUV4-20H1) and KMT5C (SUV4-20H2) mediate histone H4K20me3, whereas PHD finger protein 2 (PHF2) demethylates this residue.17, 18 NSD1 is also reported to trimethylate H4K20.9 H4K20me3 is necessary for the repressive pathway that induce pericentric heterochromatin required for G2/M arrest in response to DNA damage and recruitment of DNA repair complex.17, 19 In addition, loss of H4K20me3 is associated with telomere elongation and derepression of telomere recombination.20 Depletion of KMT5B or KMT5C leads to increased telomere elongation, which is common in cancer.20
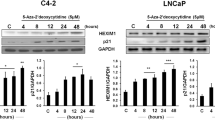
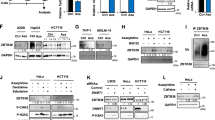
Develo** drugs that reverse cancer-specific histone modifications is one of our goals. Towards this end, we targeted the transcription factor nuclear factor-kappaB (NF-κB), which is activated in cancers and regulates inflammation-associated epigenetic changes.21, 22 Using pharmacological and genomic approaches, we show upregulation of NSD1 and SETD2 upon NF-κB inhibition. Dimethylaminoparthenolide (DMAPT), the pharmacological inhibitor of NF-κB, additionally increased KMT5C and H4K20me3 independent of its NF-κB inhibition attribute.
Results
The effects of DMAPT on histone modifications
Parthenolide, an NF-κB inhibitor, reduces histone deacetylase 1 (HDAC-1) and DNA methyltransferase 1 independent of NF-κB inhibition.23, Supplementary Table S1.
Apoptosis assay
Apoptosis was measured using Annexin V labeling using the Apoptosis Assay Kit from Invitrogen, and the number of apoptotic cells after 24 h of DMAPT treatment was measured by flow cytometry. Both floating and adherent cells were collected and stained with Alexa Fluor-488-conjugated Annexin V and propidium iodide. Annexin V-positive cells are apoptotic, propidium iodide-positive cells are necrotic and double-positive cells are necroapoptotic.
Histone extraction
Cells were lysed with Triton Extraction Buffer (TEB: PBS containing 0.5% Triton X 100(v/v), 2 mM phenylmethylsulfonylfluoride (PMSF), 0.02% (w/v) NaN3) and centrifuged at 6500 × g for 10 min at 4 °C to collect nuclei. The histones were subsequently extracted with 0.2 M HCl (Abcam histone extraction protocol, Cambridge, MA, USA).
Electrophoretic mobility gel shift assay
MDA-MB-231 and MEF cells were harvested in their exponential growth phase with or without TNFα (5 ng/ml, R&D Systems, Minneapolis, MN, USA) treatment for 15 min and assayed for NF-κB and SP-1 (as a control) DNA-binding activity as described previously.39 Antibodies for supershift assays were purchased from Santa Cruz (c-Rel, cat. no. sc-070) and Millipore (p65, cat. no. 06-418; p50, cat. no. 06-886).
Statistical analysis
Results of qRT-PCR were analyzed using the GraphPad software (www.Graphpad.com). Analysis of variance was used to determine the P-values between mean measurements. A P-value of <0.05 was deemed significant.
Analysis of public databases for prognostic relevance of NSD1 and SETD2
Expression array data of various bladder cancer stages were obtained from NCBI GEO (GDS1479), and average±S.D. was calculated. NSD1 expression data were from a single affymetrix probe available in the data set, whereas average from three probes was used for SETD2. For breast cancer, analysis of TCGA data set55 for NSD1 and SETD2 expression is presented although similar analysis using a public data set with gene expression pattern in tumors of 1809 breast cancer patients yielded similar results.44
Abbreviations
- CtBP1:
-
C-terminal-binding protein 1
- DMAPT:
-
dimethylaminoparthenolide
- GEO:
-
gene expression omnibus
- H3K27me3:
-
histone 3 lysine 27 trimethylation
- H3K36me3:
-
histone 3 lysine 36 trimethylation
- H4K20me3:
-
histone 4 lysine 20 trimethylation
- HDAC-1:
-
histone deacetylase 1
- IκBαSR:
-
inhibitor-of-kappaB alpha super-repressor
- JNK:
-
c-Jun N-terminal kinase
- KMT:
-
lysine methyltransferases
- KDM:
-
lysine demethylases
- MEF:
-
mouse embryonic fibroblast
- NF-κB:
-
nuclear factor-kappaB
- NSD1:
-
nuclear receptor-binding SET domain protein 1
- NUP98:
-
nucleoporin 98 kDa
- PARP:
-
poly-(ADP-ribose) polymerase
- PHF2:
-
PHD finger protein 2
- qRT-PCR:
-
quantitative reverse transcription-PCR
- SETD2:
-
SET domain containing 2
- TBS-T:
-
Tris-buffered saline-Tween 20
- TCGA:
-
The Cancer Genome Atlas
- TNFα:
-
tumor necrosis factor alpha
References
Sharma S, Kelly TK, Jones PA . Epigenetics in cancer. Carcinogenesis 2010; 31: 27–36.
Jones PA, Baylin SB . The epigenomics of cancer. Cell 2007; 128: 683–692.
Ben-Porath I, Thomson MW, Carey VJ, Ge R, Bell GW, Regev A et al. An embryonic stem cell-like gene expression signature in poorly differentiated aggressive human tumors. Nat Genet 2008; 40: 499–507.
Kanno R, Janakiraman H, Kanno M . Epigenetic regulator polycomb group protein complexes control cell fate and cancer. Cancer Sci 2008; 99: 1077–1084.
Schuettengruber B, Chourrout D, Vervoort M, Leblanc B, Cavalli G . Genome regulation by polycomb and trithorax proteins. Cell 2007; 128: 735–745.
Wagner EJ, Carpenter PB . Understanding the language of Lys36 methylation at histone H3. Nat Rev Mol Cell Biol 2012; 13: 115–126.
Schmidt CK, Jackson SP . On your mark, get SET(D2), go! H3K36me3 primes DNA mismatch repair. Cell 2013; 153: 513–515.
Lucio-Eterovic AK, Singh MM, Gardner JE, Veerappan CS, Rice JC, Carpenter PB . Role for the nuclear receptor-binding SET domain protein 1 (NSD1) methyltransferase in coordinating lysine 36 methylation at histone 3 with RNA polymerase II function. Proc Natl Acad Sci USA 2010; 107: 16952–16957.
Berdasco M, Ropero S, Setien F, Fraga MF, Lapunzina P, Losson R et al. Epigenetic inactivation of the Sotos overgrowth syndrome gene histone methyltransferase NSD1 in human neuroblastoma and glioma. Proc Natl Acad Sci USA 2009; 106: 21830–21835.
Weinstein JN, Lorenzi PL . Cancer: discrepancies in drug sensitivity. Nature 2013; 504: 381–383.
Mendes-Pereira AM, Sims D, Dexter T, Fenwick K, Assiotis I, Kozarewa I et al. Genome-wide functional screen identifies a compendium of genes affecting sensitivity to tamoxifen. Proc Natl Acad Sci USA 2012; 109: 2730–2735.
Wang GG, Cai L, Pasillas MP, Kamps MP . NUP98-NSD1 links H3K36 methylation to Hox-A gene activation and leukaemogenesis. Nat Cell Biol 2007; 9: 804–812.
Kuo AJ, Cheung P, Chen K, Zee BM, Kioi M, Lauring J et al. NSD2 links dimethylation of histone H3 at lysine 36 to oncogenic programming. Mol Cell 2011; 44: 609–620.
Gerlinger M, Rowan AJ, Horswell S, Larkin J, Endesfelder D, Gronroos E et al. Intratumor heterogeneity and branched evolution revealed by multiregion sequencing. N Engl J Med 2012; 366: 883–892.
Yuan W, Xu M, Huang C, Liu N, Chen S, Zhu B . H3K36 methylation antagonizes PRC2-mediated H3K27 methylation. J Biol Chem 2011; 286: 7983–7989.
Fraga MF, Ballestar E, Villar-Garea A, Boix-Chornet M, Espada J, Schotta G et al. Loss of acetylation at Lys16 and trimethylation at Lys20 of histone H4 is a common hallmark of human cancer. Nat Genet 2005; 37: 391–400.
Schotta G, Lachner M, Sarma K, Ebert A, Sengupta R, Reuter G et al. A silencing pathway to induce H3-K9 and H4-K20 trimethylation at constitutive heterochromatin. Genes Dev 2004; 18: 1251–1262.
Stender JD, Pascual G, Liu W, Kaikkonen MU, Do K, Spann NJ et al. Control of proinflammatory gene programs by regulated trimethylation and demethylation of histone H4K20. Mol Cell 2012; 48: 28–38.
Kouzarides T . Chromatin modifications and their function. Cell 2007; 128: 693–705.
Benetti R, Gonzalo S, Jaco I, Schotta G, Klatt P, Jenuwein T et al. Suv4-20h deficiency results in telomere elongation and derepression of telomere recombination. J Cell Biol 2007; 178: 925–936.
Karin M, Cao Y, Greten FR, Li ZW . NF-kappaB in cancer: from innocent bystander to major culprit. Nat Rev Cancer 2002; 2: 301–310.
De Santa F, Totaro MG, Prosperini E, Notarbartolo S, Testa G, Natoli G . The histone H3 lysine-27 demethylase Jmjd3 links inflammation to inhibition of polycomb-mediated gene silencing. Cell 2007; 130: 1083–1094.
Gopal YN, Arora TS, Van Dyke MW . Parthenolide specifically depletes histone deacetylase 1 protein and induces cell death through ataxia telangiectasia mutated. Chem Biol 2007; 14: 813–823.
Liu Z, Liu S, **e Z, Pavlovicz RE, Wu J, Chen P et al. Modulation of DNA methylation by a sesquiterpene lactone parthenolide. J Pharmacol Exp Ther 2009; 329: 505–514.
Shanmugam R, Kusumanchi P, Appaiah H, Cheng L, Crooks P, Neelakantan S et al. A water soluble parthenolide analog suppresses in vivo tumor growth of two tobacco-associated cancers, lung and bladder cancer, by targeting NF-kappaB and generating reactive oxygen species. Int J Cancer 2011; 128: 2481–2494.
Price BD, D'Andrea AD . Chromatin remodeling at DNA double-strand breaks. Cell 2013; 152: 1344–1354.
Modak R, Das Mitra S, Krishnamoorthy P, Bhat A, Banerjee A, Gowsica BR et al. Histone H3K14 and H4K8 hyperacetylation is associated with Escherichia coli-induced mastitis in mice. Epigenetics 2012; 7: 492–501.
Patel NM, Nozaki S, Shortle NH, Bhat-Nakshatri P, Newton TR, Rice S et al. Paclitaxel sensitivity of breast cancer cells with constitutively active NF-kappaB is enhanced by IkappaBalpha super-repressor and parthenolide. Oncogene 2000; 19: 4159–4169.
Hoffmann A, Leung TH, Baltimore D . Genetic analysis of NF-kappaB/Rel transcription factors defines functional specificities. EMBO J 2003; 22: 5530–5539.
Nozaki S, Sledge GW Jr, Nakshatri H . Repression of GADD153/CHOP by NF-kappaB: a possible cellular defense against endoplasmic reticulum stress-induced cell death. Oncogene 2001; 20: 2178–2185.
Mott JL, Kurita S, Cazanave SC, Bronk SF, Werneburg NW, Fernandez-Zapico ME . Transcriptional suppression of mir-29b-1/mir-29a promoter by c-Myc, hedgehog, and NF-kappaB. J Cell Biochem 2010; 110: 1155–1164.
Lin YC, Hsu EC, Ting LP . Repression of hepatitis B viral gene expression by transcription factor nuclear factor-kappaB. Cell Microbiol 2009; 11: 645–660.
Dhawan P, Richmond A . Role of CXCL1 in tumorigenesis of melanoma. J Leukoc Biol 2002; 72: 9–18.
Chua HL, Bhat-Nakshatri P, Clare SE, Morimiya A, Badve S, Nakshatri H . NF-kappaB represses E-cadherin expression and enhances epithelial to mesenchymal transition of mammary epithelial cells: potential involvement of ZEB-1 and ZEB-2. Oncogene 2007; 26: 711–724.
Kapoor-Vazirani P, Kagey JD, Vertino PM . SUV420H2-mediated H4K20 trimethylation enforces RNA polymerase II promoter-proximal pausing by blocking hMOF-dependent H4K16 acetylation. Mol Cell Biol 2011; 31: 1594–1609.
Kooistra SM, Helin K . Molecular mechanisms and potential functions of histone demethylases. Nat Rev Mol Cell Biol 2012; 13: 297–311.
Lu T, Jackson MW, Singhi AD, Kandel ES, Yang M, Zhang Y et al. Validation-based insertional mutagenesis identifies lysine demethylase FBXL11 as a negative regulator of NFkappaB. Proc Natl Acad Sci USA 2009; 106: 16339–16344.
Ge R, Wang Z, Zeng Q, Xu X, Olumi AF . F-box protein 10, an NF-kappaB-dependent anti-apoptotic protein, regulates TRAIL-induced apoptosis through modulating c-Fos/c-FLIP pathway. Cell Death Differ 2011; 18: 1184–1195.
Nakshatri H, Rice SE, Bhat-Nakshatri P . Antitumor agent parthenolide reverses resistance of breast cancer cells to tumor necrosis factor-related apoptosis-inducing ligand through sustained activation of c-Jun N-terminal kinase. Oncogene 2004; 23: 7330–7344.
Gao J, Aksoy BA, Dogrusoz U, Dresdner G, Gross B, Sumer SO et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci Signal 2013; 6: pl1.
Cerami E, Gao J, Dogrusoz U, Gross BE, Sumer SO, Aksoy BA et al. The cBio cancer genomics portal: an open platform for exploring multidimensional cancer genomics data. Cancer Discov 2012; 2: 401–404.
Dyrskjot L, Kruhoffer M, Thykjaer T, Marcussen N, Jensen JL, Moller K et al. Gene expression in the urinary bladder: a common carcinoma in situ gene expression signature exists disregarding histopathological classification. Cancer Res 2004; 64: 4040–4048.
Goswami CP, Nakshatri H . PROGgene: gene expression based survival analysis web application for multiple cancers. J Clin Bioinformatics 2013; 3: 22.
Gyorffy B, Lanczky A, Eklund AC, Denkert C, Budczies J, Li Q et al. An online survival analysis tool to rapidly assess the effect of 22,277 genes on breast cancer prognosis using microarray data of 1,809 patients. Breast Cancer Res Treat 2010; 123: 725–731.
Nakshatri H, Bhat-Nakshatri P, Martin DA, Goulet RJ Jr, Sledge GW Jr . Constitutive activation of NF-kappaB during progression of breast cancer to hormone-independent growth. Mol Cell Biol 1997; 17: 3629–3639.
Zhou Y, Eppenberger-Castori S, Eppenberger U, Benz CC . The NFkappaB pathway and endocrine-resistant breast cancer. Endocr Relat Cancer 2005; 12: S37–S46.
Riggins RB, Zwart A, Nehra R, Clarke R . The nuclear factor kappa B inhibitor parthenolide restores ICI 182,780 (Faslodex; fulvestrant)-induced apoptosis in antiestrogen-resistant breast cancer cells. Mol Cancer Ther 2005; 4: 33–41.
Czabotar PE, Lessene G, Strasser A, Adams JM . Control of apoptosis by the BCL-2 protein family: implications for physiology and therapy. Nat Rev Mol Cell Biol 2013; 15: 49–63.
Huang N, vom Baur E, Garnier JM, Lerouge T, Vonesch JL, Lutz Y et al. Two distinct nuclear receptor interaction domains in NSD1, a novel SET protein that exhibits characteristics of both corepressors and coactivators. EMBO J 1998; 17: 3398–3412.
Stephens PJ, Tarpey PS, Davies H, Van Loo P, Greenman C, Wedge DC et al. The landscape of cancer genes and mutational processes in breast cancer. Nature 2012; 486: 400–404.
**e P, Tian C, An L, Nie J, Lu K, **ng G et al. Histone methyltransferase protein SETD2 interacts with p53 and selectively regulates its downstream genes. Cell Signal 2008; 20: 1671–1678.
Dong J, Jimi E, Zhong H, Hayden MS, Ghosh S . Repression of gene expression by unphosphorylated NF-kappaB p65 through epigenetic mechanisms. Genes Dev 2008; 22: 1159–1173.
Beauchef G, Bigot N, Kypriotou M, Renard E, Poree B, Widom R et al. The p65 subunit of NF-kappaB inhibits COL1A1 gene transcription in human dermal and scleroderma fibroblasts through its recruitment on promoter by protein interaction with transcriptional activators (c-Krox, Sp1, and Sp3). J Biol Chem 2012; 287: 3462–3478.
Beg AA, Baltimore D . An essential role for NF-kappaB in preventing TNF-alpha-induced cell death [see comments]. Science 1996; 274: 782–784.
Koboldt DC, Fulton RS, McLellan MD, Schmidt H, Kalicki-Veizer J, McMichael JF et al. Comprehensive molecular portraits of human breast tumours. Nature 2012; 490: 61–70.
Acknowledgements
We thank Dr. Alex Hoffman for MEFs and Nikail R Collins for technical assistance. This work is supported by a grant from NIH CA143994-01A1 to HN. IUPUI Breast Cancer Signature Center initiative provided funding for establishing prognostic database.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
HN, PAC, WM, and CS are the co-founders of Leuchemix Inc., which is develo** parthenolide and its analogs as anti-cancer agents. The other authors declare no conflict of interest.
Additional information
Edited by D Heery
Supplementary Information accompanies this paper on Cell Death and Disease website
Rights and permissions
Cell Death and Disease is an open-access journal published by Nature Publishing Group. This work is licensed under a Creative Commons Attribution 4.0 International Licence. The images or other third party material in this article are included in the article’s Creative Commons licence, unless indicated otherwise in the credit line; if the material is not included under the Creative Commons licence, users will need to obtain permission from the licence holder to reproduce the material. To view a copy of this licence, visit http://creativecommons.org/licenses/by/4.0
About this article
Cite this article
Nakshatri, H., Appaiah, H., Anjanappa, M. et al. NF-κB-dependent and -independent epigenetic modulation using the novel anti-cancer agent DMAPT. Cell Death Dis 6, e1608 (2015). https://doi.org/10.1038/cddis.2014.569
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/cddis.2014.569
- Springer Nature Limited
This article is cited by
-
Bioactivities and the structural modification of Parthenolide: a review
Medicinal Chemistry Research (2024)
-
Connectivity map** of glomerular proteins identifies dimethylaminoparthenolide as a new inhibitor of diabetic kidney disease
Scientific Reports (2020)
-
Parthenolide promotes the repair of spinal cord injury by modulating M1/M2 polarization via the NF-κB and STAT 1/3 signaling pathway
Cell Death Discovery (2020)
-
NF-κB, inflammation, immunity and cancer: coming of age
Nature Reviews Immunology (2018)
-
NF-κB inhibition by dimethylaminoparthenolide radiosensitizes non-small-cell lung carcinoma by blocking DNA double-strand break repair
Cell Death Discovery (2018)