Abstract
Tumor necrosis factor-α-induced protein 8 (TNFAIP8) is a stress-response gene that has been associated with cancer, but no studies have differentiated among or defined the regulation or function of any of its several recently described expression variants. We found that TNFAIP8 variant 2 (v2) is overexpressed in multiple human cancers, whereas other variants are commonly downregulated in cancer (v1) or minimally expressed in cancer or normal tissue (v3–v6). Silencing v2 in cancer cells induces p53-independent inhibition of DNA synthesis, widespread binding of p53, and induction of target genes and p53-dependent cell cycle arrest and DNA damage sensitization. Cell cycle arrest induced by v2 silencing requires p53-dependent induction of p21. In response to the chemotherapeutic agent doxorubicin, p53 regulates v2 through binding to an intragenic enhancer, together indicating that p53 and v2 engage in complex reciprocal regulation. We propose that TNFAIP8 v2 promotes human cancer by broadly repressing p53 function, in essence offsetting p53-dependent tumor suppression.
Similar content being viewed by others
Main
Tumor necrosis factor-α-induced protein 8 (TNFAIP8) is the founding member of a recently described family of environmental stress response genes that are induced by TNFα. TNFAIP8 has been shown to promote or inhibit apoptosis, depending on cell type and context.1, 2 Although little is known about TNFAIP8, it has recently been found to be overexpressed in a wide range of human cancers. Some studies have suggested protumor functions for TNFAIP8, including enhancement of cell survival, proliferation, and metastasis3, 4, 5, 6, 7, 8, 9 and resistance to cancer chemotherapeutics in mice.4, 10 Nonetheless, nothing is known about how TNFAIP8 influences responses to DNA damage in cancer cells. Moreover, several transcript variants from the TNFAIP8 gene were recently registered in the NCBI reference sequence database, but no study to date has differentiated among them or defined the factors that govern their expression.
The tumor suppressor p53 is a transcription factor that regulates many biological processes through its target gene network. Some of the well-characterized p53 target genes including p21/CDKN1A, growth arrest and DNA damage-inducible 45a (GADD45A) and Bcl-2-like protein 4 (BAX) together promote senescence, cell cycle arrest, and apoptosis, all of which may contribute to the tumor suppressing functions of p53.11, 12 Additional noncanonical tumor-suppressive pathways from p53 have recently been identified.13, 14, 15 Improved characterization of the p53 DNA-binding sequence with modern sequencing techniques has led to the identification of a rapidly expanding list of p53 target genes.16, 17, 18 Continued identification of these genes and characterization of their mechanisms of regulation and function will be critical to a full understanding of p53 and for full realization of opportunities to intervene upon p53 in cancer.
Here, we identify TNFAIP8 variant 2 (v2) as a p53-regulated gene product that promotes cancer through complex, reciprocal regulatory interactions with p53. We show that TNFAIP8 v2 may contribute to both carcinogenesis and chemotherapeutic resistance by broadly suppressing p53 activity, thus offsetting p53-dependent tumor suppression.
Results
TNFAIP8 v1 and v2 are differentially expressed in human cancers
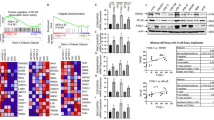
Recent revisions of the human reference sequence database indicate that TNFAIP8 (chr5: 118 604 418–118 730 138) has six expression (mRNA) variants. The last exon is common, whereas usage of other exons differs across variants (Supplementary Figure S1). Several reports have shown that TNFAIP8 is overexpressed in human cancers,3, 4, 5, 8, 19, Full size image
Having identified a p53-binding site, we further characterized p53 regulation of TNFAIP8 using U2OS cells as a model system. We cloned a ~600 bp region encompassing the p53RE upstream of a luciferase reporter. An empty vector, the v2 p53RE vector, or a positive control vector with confirmed functional p53REs, each upstream of a luciferase reporter construct, were individually transfected into U2OS cells stably expressing either a scrambled or p53-directed shRNA (Figure 3c). Treatment with DOX significantly enhanced luciferase activity (v2 p53RE and p53+ only) in scramble shRNA cells. Both basal and DOX-induced luciferase were significantly reduced in p53-depleted cells. These results confirm that the p53RE in the v2 intron is functional as this region confers p53-dependent transactivation ability.
The intronic p53-binding region in TNFAIP8 is an enhancer
The p53-binding region lies within the first intron of v2. However, it is ~50 kb downstream from the v2 promoter region and ~30 kb upstream from the v1 promoter region, suggesting that it may possibly be an intragenic enhancer. To address this possibility, we analyzed it for enhancer-like chromatin marks. Chromatin marks characteristic of enhancers include high histone H3 mono-methylation on lysine 4 (‘H3K4me1’), high histone H3 acetylation on lysine 27 (‘H3K27ac’) and low histone H3 tri-methylation on lysine 4 (‘H3K4me3’). These contrast with characteristic promoter chromatin marks (low H3K4me1, high H3K27ac, high H3K4me3).25 Using ChIP-PCR, we measured these marks in U2OS cells in the intronic p53-binding region and, for comparison purposes, in the v2 promoter region (V2P). As shown in Figure 3d, regardless of DOX treatment, v2 p53RE has significantly higher H3K4me1 and lower H3K4me3 compared with V2P. Both sites show moderate levels of H3K27ac. Thus, the v2 p53RE region has marks characteristic of an enhancer.
To further validate v2 p53RE as an intragenic enhancer, three-dimensional chromatin loo** assays using EcoRI restriction of fixed chromatin followed by PCR using primers designed to survey the region were performed to test whether v2 p53RE physically interacts with V2P despite being ~50 kb removed. Activated p53 has been shown to act on preexisting chromatin loops.25 The primer pair ‘1/4’ yielded significant amplification (Figure 3e), indicating ligation between restricted ends of sites 1 and 4 and the physical interaction between the v2 promoter and the v2 p53RE region through chromatin loo**. These data, together with the presence of enhancer-like chromatin marks and transactivation capacity of v2 p53RE, suggest that p53 regulates DOX-induced v2 expression through binding to the v2 p53RE intragenic enhancer region.
TNFAIP8 v2 silencing induces widespread p53 binding and p53 target induction
Given our finding that v2 is upregulated across several human cancers and that basal v2 expression in cancer cell lines is p53 independent, we hypothesized that v2 might promote cancer cell survival through suppressing p53 activities. Several prior examples of reciprocal p53 target regulatory loops have been described.26 In order to address this, we transiently expressed TNFAIP8-directed shRNA (‘TP8i’) versus scramble shRNA (‘scri’) in (v1-null) A549 cells (Figure 4a). Unexpectedly, we found that p53 protein levels in nuclear extracts and whole-cell lysates were decreased in v2-depleted cells (Figure 4b). We reasoned that one possible explanation might be that p53 was relocalized to chromatin and therefore insoluble in the extraction buffer. To address this, we isolated chromatin-bound nuclear protein and, using specific antibodies, measured both total p53 and acetylated p53, as acetylation is associated with p53 activation and enhanced binding to p53REs.27 Both total and acetylated p53 were indeed increased in the chromatin-bound nuclear fraction in v2-depleted cells, suggesting that v2 silencing regulates p53 by promoting its acetylation and localization to chromatin (Figure 4c). ChIP assays also revealed that p53 enrichment at the promoter regions of multiple established p53 target genes was significantly greater in v2-depleted cells than in control cells (Figure 4d).
TNFAIP8 v2 depletion induces p53 binding and p53 target expression. (a) Immunoblot of v2 and actin (control) in whole-cell lysates of A549 cells treated with TNFAIP8 (TP8i) or scrambled (scri) shRNA. (b) Immunoblot of p53 from nuclear and whole-cell extracts of cells under indicated treatments. (c) Immunoblot of acetylated p53-K382 (‘p53-ac’) and total p53 in chromatin-bound nuclear extracts of p53+ or p53-silenced A549 cells treated with scri or TP8i. Histone 3 (‘H3’) and actin are loading controls. (d) p53 ChIP-PCR of promoter regions of indicated target genes in TP8i and scri cells. IgG serves as negative control. (e and f) mRNA expression of p21 and Gadd45a (RT-PCR) (e) and FAS and 14-3-3σ (Nanostring) (f) was measured in p53+ or p53-silenced A549 cells treated with scri or TP8i. (g) v2 mRNA expression (normalized to GusB) and (h) p53 binding to the indicated target genes was measured with RT-PCR and ChIP-PCR, respectively, in A549 cells transiently transfected with an empty vector (‘empty’) or a v2 expression vector (‘v2’). Results are representative of three or more independent experiments. *P<0.05
We verified that some target genes displaying increased p53 binding also had increased expression in TP8i cells (Figures 4e and f). A second TNFAIP8-directed shRNA, which depleted v2 by ~37%, resulted in similar expression changes (Supplementary Figures S3A and B). Not all genes displaying increased p53 binding upon v2 silencing had a corresponding change in mRNA expression (e.g., MDM2, DDB2; data not shown). This is consistent with past reports that mRNA expression of p53 target genes may be regulated by mechanisms beyond binding.17 As v2 depletion led to enhanced p53 activity, we predicted that overexpression of v2 may inhibit p53 activity. v2 overexpression indeed decreased p53 binding to several targets (Figures 4g and h). Collectively, these findings suggest that TNFAIP8 v2 exerts complex regulation of p53 function in cancer cells, where it restrains p53 from activating select nuclear genes.
TNFAIP8 v2 depletion inhibits DNA synthesis and leads to cell cycle arrest
To date, no study has defined variant-specific functions for TNFAIP8 in cancer. As we found that v2 was overexpressed in cancers and appears to suppress p53, we hypothesized that v2 might promote cancer cell growth. To address this, v2 silencing was performed in A549 cells stably expressing either p53-directed shRNA (‘p53i’) or a scramble shRNA (‘p53+’)(Figure 5a) and DNA synthesis examined using 5-bromo-2’-deoxyuridine (BrdU) incorporation. As shown in Figure 5b, p53+ and p53i cells with scramble shRNA displayed considerable BrdU staining intensity. In stark contrast, v2-depleted p53+ cells showed low BrdU staining. Quantification of BrdU intensity revealed a significant difference between TP8i and control cells in both the p53+ and p53i background (Figure 5c). As the effect of v2 depletion on DNA synthesis was less in p53i than p53+ cells (37% versus 68% reduction; P<0.05), we conclude that DNA synthesis arrest in TP8i cells is at least partially p53 dependent, but can nonetheless proceed in p53-deficient cells. Interestingly, there was no significant difference in the percentage of TP8i and Scri cells that were BrdU positive (Supplementary Figure S3C), indicating that DNA synthesis is initiated in v2-depleted cells but unable to progress.
TNFAIP8 v2 depletion stalls DNA replication and induces p53-dependent cell cycle arrest. (a) TNFAIP8 v2 mRNA levels were quantified in p53+ and p53-silenced (p53i) A549 cells untransduced (‘−’) or transduced with scramble (‘scri’) or TNFAIP8-directed (‘TP8i’) shRNA. At right, an immunoblot of v2 under the same conditions is shown. (b) 5-Bromo-2’-deoxyuridine (BrdU) incorporation and 7AAD staining were quantified using flow cytometry in p53+ and p53i A549 cells transduced with scri or TP8i shRNA. BrdU+ cells (S phase), yellow; G1, green; G2, purple. (c) Quantification of BrdU intensity of cells in (b). (d) Cell cycle profile based on 7AAD staining of cells in (a). G2-phase cells (purple) are defined as cells with double the nuclear 7AAD stain intensity compared with G1-phase cells (green). S-phase cells are BrdU+ (yellow). (e and f) mRNA levels of PCNA (RT-PCR) (e) and cyclins D1, E1, and E2 (Nanostring) (f) of cells under indicated treatments. (g) p21 quantified by RT-PCR and immunoblot in p53+ A549 cells transduced with either scri or TP8i shRNA, followed by transfection of scramble (‘scri’) or p21 (‘p21i’) siRNA. (h) Cell cycle analysis of cells described in (g). Results are representative of three or more independent experiments. *P<0.05
Given that v2-depleted cells displayed defective DNA synthesis, we next analyzed the cell cycle profile. Cell cycle analysis using 7-aminoactinomycin D (7AAD) revealed a stalling in S phase of v2-depleted cells compared with controls (Figure 5d and Supplementary Figure S3D). Despite our finding that the DNA synthesis defect caused by v2 depletion occurs in both p53+ and p53i cells, v2-depleted p53i cells did not arrest in the S phase, indicating a requirement for p53 in the S-phase arrest (Figure 5d).
GADD45A and CDKN1A (p21), both of which regulate cell cycle arrest28, 29 and are induced by p53, were significantly upregulated in v2-depleted cells compared with controls in a p53+, but not p53i, background (Figure 4d and Supplementary Figure S3B), suggesting they may contribute to the p53-dependent S-phase arrest in v2-depleted cells. On the other hand, proliferating cell nuclear antigen (PCNA), which is required for progression of DNA replication,30 was significantly reduced in TP8i cells in both the p53+ and p53i settings (Figure 5e). As PCNA is dispensable for initiation of DNA replication but required for progression,30 our finding that PCNA is decreased in v2-depleted cells irrespective of p53 status is consistent with our finding of a defect in the progression but not initiation of DNA synthesis in both p53+ and p53i v2-depleted cells (Figures 5b and c).
Cyclins D1, E1, and E2 are all required for transition from G1 to S phase, after which the expression of these cyclins must be decreased to maintain the proliferation signal and progress through S phase.31, 32, 33 We hypothesized that expression of these cyclins might be increased in v2-depleted cells given the observed S-phase arrest (Figure 5d). Consistent with our prediction, cyclins D1, E1, and E2 were all significantly increased in v2-depleted cells (Figure 5f). Unlike the case for cyclins E1 and E2, cyclin D1 induction in TP8i cells appeared to be p53 dependent. This finding is consistent with previous reports describing p53 enhancement of cyclin D1 expression and subsequent growth arrest.34, 35
To test whether the S-phase arrest induced by v2 depletion was specific for A549 cells, we performed similar experiments in U2OS, MCF-7 (breast cancer), and HCT116 cells, all of which, like A549 cells, predominantly express v2 and little v1. As with A549 cells, v2 depletion in all cell lines significantly upregulated p21 (Supplementary Figures S4A–C) and impaired proliferation (not depicted). However, the percentage of cells in G1 was significantly increased in v2-depleted U2OS and MCF-7 cells, whereas v2-depleted HCT116 cells exhibited an increase in G2/M, indicating an arrest in G1 and G2/M, respectively (Supplementary Figures S4D–F and S5). Taken together, all cancer cell lines surveyed displayed upregulation of p21 with v2 depletion, but, interestingly, exhibited a variety of types of cell cycle arrest.
Cell cycle arrest in v2-depleted cells is primarily due to p21
Our results in A549 cells indicated that v2 depletion leads to defective DNA replication progression, potentially through downregulated PCNA and S-phase stalling. However, in an effort to identify gene networks in a more unbiased manner that might account for these findings, gene expression was profiled in A549 cells using Nanostring technology. Of 770 genes in the Nanostring PanCancer Codeset, 221 (28.7%) were significantly altered in expression in response to v2 silencing. Ingenuity Pathway Analysis revealed that the highest scoring network of genes with significant expression changes upon v2 silencing in p53+ cells was that associated with cell proliferation and cell cycle, and that this network centered upon CDKN1A (i.e., p21) (Supplementary Figure S6). In order to address whether p21 orchestrates the p53-dependent cell cycle arrest induced by v2 depletion, we introduced scrambled or pooled p21-directed siRNA into TP8i and scri, p53+ A549 cells (Figure 5g). p21 silencing prevented the S-phase arrest in v2-depleted cells (Figure 5h), confirming p21 as a major p53-responsive driver of the defective cell growth of TP8i cells.
TNFAIP8 v2 depletion enhances damage-induced apoptosis
We next evaluated the impact of v2 on the response of cancer cells to chemotherapeutic challenge. As shown in Figures 6a and b, DOX induced a dramatic, nearly ninefold higher frequency of apoptosis (51% versus 6%) in v2-depleted cells than in controls. The increased apoptosis appears to be at least partially p53 dependent (Figure 6b). Similar results were seen with the nongenotoxic agent staurosporine36 (Figure 6c and Supplementary Figure S7). TP8i cells also showed significantly higher activation of the executioner caspases 3/7 in response to both DOX and staurosporine in a p53+, but not p53i, background (Figure 6d). Depletion of v2 thus sensitizes cancer cells to p53-dependent DOX- and staurosporine-induced apoptosis. These results suggest that TNFAIP8 v2 is an anti-apoptotic regulator in response to chemotherapeutics, attenuating chemotherapeutic efficacy by reducing p53-dependent death.
TNFAIP8 v2 depletion enhances damage-induced apoptosis. (a) Annexin V and propidium iodide (PI) staining were quantified by flow cytometry in p53+ and p53i A549 cells transduced with a scramble (‘Scri’) or TNFAIP8-directed (‘TP8i’) shRNA and untreated or treated with DOX. (b and c) The percentage of Annexin V+ cells was quantified in untreated and DOX-treated cells (b), and in DMSO-treated cells (vehicle control) and staurosporine (‘Stauro’)-treated cells (c). (d) The fold change in caspase 3/7 activity is shown in the cells described in (b and c) after DOX or Stauro treatment. DOX and Stauro values are normalized to untreated or DMSO-treated values, respectively. Results are representative of three or more independent experiments. *P<0.05