Abstract
Background:
The FAST was a factorial trial in first-line treatment of advanced non-small-cell lung cancer (NSCLC), addressing the role of replacing cisplatin with a non-platinum agent. The prognostic and predictive effect of ERCC1/BRCA1 expression and ERCC1/XPD/XRCC1–3 gene polymorphisms on outcomes of patients was examined.
Methods:
Patients were randomised to receive treatment with or without cisplatin. ERCC1/BRCA1 expression was determined by immunohistochemistry. ERCC1 (C8092A, C118T), XPD (Lys751Gln), XRCC1 (Arg399Gln) and XRCC3 (Thr241Met) gene polymorphisms were evaluated on tumour DNA by TaqMan allelic discrimination assay.
Results:
Tumour samples were available from 110 of 433 patients enrolled: 54.7% were ERCC1 positive and 51.4% were BRCA1 positive. Overall, ERCC1-negative patients had better response rate (P=0.004), progression-free survival (P=0.023) and overall survival (P=0.012) compared with positive ones, with no statistically significant treatment interaction. The BRCA1-positive patients showed numerically better outcomes, although not statistically significant, with no treatment interaction. Among DNA repair gene polymorphisms, only XRCC1 Gln/Gln genotype evidenced a potential prognostic role (P=0.036).
Conclusion:
This study confirms the prognostic role of ERCC1 expression and XRCC1 (Arg399Gln) polymorphism in advanced NSCLC treated with first-line chemotherapy. None of these biomarkers was shown to be a specific predictive factor of cisplatin efficacy.
Similar content being viewed by others
Main
With the exception of EGFR mutated tumours, the current standard of care in first-line treatment of advanced non-small-cell lung cancer (NSCLC) is based on a combination of cisplatin associated with a third-generation chemotherapeutic agent (Azzoli et al, 2009; Peters et al, 2012).
The mechanism of action of platinum agents is based on the formation of DNA adducts that results in the alteration of DNA structure and, eventually, inhibition of DNA replication and transcription (Martin et al, 2008). Hence, the expression of genes involved in mechanisms of DNA repair has been studied as possible predictive factor in patients treated with platinum-based chemotherapy.
The ERCC1 (excision repair cross-complementing 1) is a gene encoding a protein of the nucleotide excision repair (NER) complex, a group of proteins able to correct DNA damage induced by substances forming adducts, like platinum (Martin et al, 2008). High levels of ERCC1 protein have been shown to be a positive prognostic factor in chemotherapy-naive radically resected NSCLC (Olaussen et al, 2006; Zheng et al, 2007; Simon et al, 2008). On the contrary, they have been evidenced to be a negative predictive factor in patients treated with platinum chemotherapy, in both adjuvant and metastatic setting, probably because of an association between ERCC1 expression and platinum resistance as shown in NSCLC, ovarian and gastroenteric tumours (Li et al, 2000; Joshi et al, 2005; Gazdar, 2007). However, whether ERCC1 is simply a generic prognostic factor or a specific predictor of cisplatin efficacy is still to be demonstrated.
The BRCA1 (breast cancer 1) is an oncosuppressor gene involved in two kinds of DNA repair mechanisms: NER and double-strand break repair (DSBR) (Martin et al, 2008). Overexpression of BRCA1 represents a marker of platinum resistance in several cellular lines (Husain et al, 1998). Moreover, several studies have shown a possible role of BRCA1 as a predictor of response in advanced NSCLC, suggesting that mRNA expression levels of BRCA1 could be used to choose the more appropriate chemotherapy treatment, particularly with cisplatin or antimicrotubule agents (Taron et al, 2004; Reguart et al, 2008). Finally, Rosell et al (2007) have described the association between expression levels of BRCA1 and ERCC1 and have confirmed the role of BRCA1 as a prognostic factor in resected NSCLC.
In the past years, the role of single-nucleotide polymorphisms (SNPs) of ERCC1, XPD (xeroderma pigmentosum group D, also known as ERCC2), XRCC1 (X-ray cross–complementing group 1) and XRCC3 (X-ray cross–complementing group 3) genes as predictive factors for outcome in platinum chemotherapy-treated NSCLC has been thoroughly investigated (Gurubhagavatula et al, 2004; Isla et al, 2004; Ryu et al, 2004; Camps et al, 2006; de las Penas et al, 2006; Kalikaki et al, 2009; Horgan et al, 2011; Yin et al, 2011; Wei et al, 2011). The XPD gene is a member of NER pathway and encodes a protein involved in DNA transcription, whereas XRCC1 and XRCC3 are essential for base excision repair (BER) process. Data emerging from previous studies allowed to confirm that SNPs of these genes are expression of an interindividual variability in DNA repairing ability and represent possible predictive factors of response to platinum-based chemotherapy.
However, the results available in literature concerning ERCC1/BRCA1 expression and ERCC1/XPD/XRCC1-3 gene polymorphisms are, overall, not fully consistent, mainly based on retrospective single-arm studies and limited to cisplatin-based chemotherapy.
The aim of our study was to investigate the predictive role of ERCC1/BRCA1 immunohistochemical expressions and of ERCC1 C8092A and C118T, XPD Lys751Gln, XRCC1 Arg399Gln and XRCC3 Thr241Met SNPs in the context of a prospective randomised study assessing the role of platinum in advanced NSCLC (FAST trial) (Boni et al, 2012).
Materials and methods
Patients
From October 2001 until July 2006, 433 patients were enrolled in multicentric factorial clinical trial FAST (Boni et al, 2012). The principal inclusion criteria were: cytological or histological diagnosis of NSCLC in locally advanced or metastatic stage; no previous chemotherapy; age ⩾18 years and performance status (PS) ⩽2. Patients were assigned to one of the four treatment arms: gemcitabine+cisplatin (GP), gemcitabine+vinorelbine (GN), gemcitabine+cisplatin+ifosfamide (GIP) or gemcitabine+vinorelbine+ifosfamide (GIN). Chemotherapy cycles were administered every 3 weeks for a maximum of six cycles; tumour response was assessed by computed tomography scan after every three cycles according to the RECIST criteria (version 1.0) (Therasse et al, 2000).
According to ancillary biological study protocol (Bio-FAST), it was planned to collect all available histological paraffin-embedded tumour samples of treated patients in order to assess ERCC1 and BRCA1 immunohistochemical expression and SNPs of ERCC1 C8092A and C118T, XPD Lys751Gln, XRCC1 Arg399Gln and XRCC3 Thr241Met.
Immunohistochemistry (IHC)
Tumour sections were incubated with specific monoclonal antibodies directed against ERCC1 epitope (clone 8F1, 1 : 200 dilution, Neomarkers, Lab Vision Corporation, Fremont, CA, USA) and BRCA1 epitope (clone MS110, 1 : 300 dilution, Calbiochem, EMD Corporation, Billerica, MA, USA). For determination of the ERCC1 status, for each sample a semiquantitative H score obtained by multiplying the intensity score with the positive nuclei proportion score was calculated, according to Olaussen et al (2006). The median value of all H scores was used as a cutoff in order to classify tumours as ERCC1 positive or negative. With regard to BRCA1 expression, only the percentage of positive nuclei was valuated and 10% was chosen as the cutoff value distinguishing between positive and negative (Wachters et al, 2005). For immunohistochemical methods see Supplementary File.
DNA isolation and genoty**
DNA was obtained from paraffin-embedded tissue and extracted using the commercial DNeasy tissue kit (Qiagen, Valencia, CA, USA) according to the manufacturer’s instructions.
The SNPs selected for this study were: ERCC1 8092C>A (rs3212986), ERCC1 C118T (rs11615), ERCC2/XPD Lys751Gln (rs13181), XRCC1 Arg399Gln (rs25487) and XRCC3 Thr241Met (rs861539). The analysis of SNPs was performed using a real-time PCR allelic discrimination TaqMan assay (Applied Biosystems, Foster City, CA, USA). For more details see Supplementary File.
Statistical methods
The distributions of all studied patients were reported with respect to their demographic, clinical and biological characteristics and were summarised as frequencies and percentages. Comparisons of proportions were performed with the χ2 test for heterogeneity or Fisher’s exact test, when appropriate. Mean ranks were compared by means of Wilcoxon two-sample test. All candidate prognostic and predictive factors were investigated for their impact on objective response rate (RR), duration of progression-free survival (PFS), defined as the interval between randomisation and disease progression or death for any cause, and overall survival (OS), defined as the time between randomisation and death, whatever the cause. Observation time of patients not progressed and alive at the last follow-up visit was censored. Confidence intervals (CIs) of median survival times were calculated according to the log-log method of Brookmeyer and Crowley. Survival rates were plotted after adjustment by treatment type, according to the corrected group prognosis method. As estimates of prognostic effect, odds ratios (ORs) and hazard ratios (HRs) with a 95% CIs were calculated with logistic regression and Cox proportional hazard models, respectively. The presence of heterogeneity of treatment effect (platinum vs non-platinum) between the strata identified by each investigated biomarker was tested introducing the appropriate interaction term into the statistical models. All estimates of ORs and HRs and their 95% CIs were obtained after adjustment by treatment type. Wald test was used to quantify the statistical significance of all coefficients. All reported P-values are two sided and significant level was set at 0.05. Statistical analyses were performed by LB at Istituto Toscano Tumori using SAS System 9.2 (SAS Corporation, Cary, NC, USA).
In the Bio-FAST study, only the comparison between platinum (GP and GIP) and non-platinum (GN and GIN), and not between two and three drugs, was considered. In two groups (platinum and non-platinum), the patient distribution according to two or three drugs was well balanced; in fact, 45% of patients treated with platinum and 43% treated with non-platinum received two drugs.
Results
Patients’ characteristics
Sufficient tumour tissue for biological assessments was obtained from 110 patients, whose characteristics are reported in Table 1. Histogical specimens were obtained either by endoscopic sampling or after surgical resection. In both cases, tissues used for our purpose were collected from the primary lung lesions.
Overall, there were no statistical differences between patients included in the Bio-FAST protocol and patients not studied for biological characteristics enrolled in the FAST trial (Table 1).
We observed partial or complete response in 35 patients (RR=31.8%); 22 of these were treated with platinum and 13 were treated without platinum. The RR was 38.6% in the platinum arm and 24.5% in the non-platinum arm (P=0.114). Four patients were alive without disease progression at the time of the analysis, with a minimum duration of follow-up of 18 months. The median PFS and OS were 6.3 (95% CI 4.3–7.1; 108 events) and 12.4 (95% CI 9.4–14.5; 106 events) months, respectively. In the platinum arm, median PFS and OS reached 6.6 (95% CI 4.8–7.3) and 11.2 (95% CI 7.1–15.7) months, respectively, vs 4.4 (95% CI 3.6–7.3) and 12.5 (95% CI 9.4–15.6) months in the arm without platinum (P=0.399 and=0.698, for PFS and OS, respectively). Concerning toxicity, no statistical differences between treatment arms were found (data not shown).
No statistically significant differences were observed in outcome obtained in Bio-FAST patients and in the population of the FAST trial not studied for biological characteristics (data not shown).
ERCC1 IHC expression
After immunohistochemical assessment, 58 (54.7%) patients resulted ERCC1 positive (data missing from 4 patients because of sample inadequacy). The characteristics of the patients according to the ERCC1 expression are reported in Supplementary Table S1.
Overall, RR was 47.9% in ERCC1-negative patients vs 19% in ERCC1-positive ones (OR 0.27; 95% CI 0.11–0.66; P=0.004). In Table 2 RRs according to the ERCC1 expression in platinum vs non-platinum treatment are reported. A trend towards a qualitative interaction was evidenced between ERCC1 expression and treatment type (P=0.071).
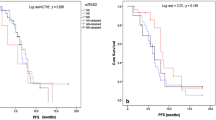
The ERCC1-negative patients had a median PFS of 7.2 (95% CI 6.5–9.1) months, whereas ERCC1-positive patients reached a median PFS of 4.3 months (95% CI 3.8–6.3; HR 1.61; 95% CI 1.07–2.44; P=0.023; Figure 1A). Median PFS values according to the ERCC1 expression in platinum vs non-platinum treatment are reported in Table 2 and in Figure 1C (P=0.984, interaction test).
Adjusted estimates of progression-free survival and overall survival in ERCC1-negative vs ERCC1-positive patients ( A and B , respectively) and also according to the treatment received, platinum vs non-platinum regimens ( C and D , respectively). The numbers of patients at risk are reported according to the distributions of the observation times. CI, confidence interval; ERCC1+, ERCC1 positive; ERCC1−, ERCC1 negative; HR, hazard ratio; P−, non-platinum; P+, platinum.
The ERCC1-negative patients showed a statistically significant longer OS compared with ERCC1-positive patients. The ERCC1-negative patients had a median OS of 16.4 (95% CI 12.6–23.9) vs 8.5 (95% CI 6.3–11.3) months reached by ERCC1-positive patients (HR 1.70; 95% CI 1.12–2.58; P=0.012; Figure 1B). Median OS values according to the ERCC1 expression in platinum vs non-platinum treatment are reported in Table 2 and in Figure 1D (P=0.446, interaction test).
BRCA1 IHC expression
Regarding BRCA1, 56 (51.4%) patients resulted positive (data missing from 1 patient because of samples inadequacy). The characteristics of the patients according to the BRCA1 expression are reported in Supplementary Table S1.
There was no statistically significant difference in RR between positive and negative patients; positive patients showed a RR of 35.7% vs 26.4% in negative ones (OR 1.69; 95% CI 0.72–3.98; P=0.231).
There was also no statistically significant difference between positive and negative patients in median PFS (HR 0.76; 95% CI 0.52–1.12; P=0.167): positive patients had a median PFS of 6.5 (95% CI 4.4–7.7) vs 5.8 (95% CI 3.8–7.0) months in negative ones.
No statistical difference was also found between BRCA1-positive and BRCA1-negative patients in OS analysis; negative patients had a median OS of 11.2 (95% CI 6.9–13.9) vs 13.5 (95% CI 9.4–17.9) months obtained in positive patients (HR 0.71; 95% CI 0.47–1.06; P=0.093).
Considering treatment arms, no differences were evidenced between BRCA1-negative and BRCA1-positive patients for all outcome variables (Table 2).
DNA repair gene polymorphisms
Genoty** was successfully performed in the vast majority of DNA samples. No discrepancies were observed in the samples analysed in duplicate (∼10%), and all the genoty** data were included in the final analysis. All polymorphisms studied followed Hardy–Weinberg equilibrium and genotype frequencies were comparable with those reported in Caucasian populations.
In Supplementary File and Supplementary Tables S2 and S3, the frequencies of all SNPs are reported. Correlation analyses of SNPs were performed grou** patients with wild-type genotype vs patients with heterozygote or homozygote variant. Overall, no significant difference emerged in RR, PFS and OS between different genotypes for all polymorphisms, except for XRCC1. In fact, patients with Gln/Gln genotype obtained a median OS of 20.4 (95% CI 7.8–38.7) months, whereas patients with Arg/Arg or Arg/Gln (Arg/−) genotype obtained a median OS of 11.3 (95% CI 8–15.4) months (HR 0.47; 95% CI 0.23–0.95; P=0.036). For more details see Supplementary File.
In Tables 3 and 4, RR, PFS and OS according to the different genotypes in platinum vs non-platinum treatment are reported.
Discussion
The two major components of NER and homologous recombination repair (HRR) pathways, ERCC1 and BRCA1 respectively, have been studied as predictive markers in the treatment of NSCLC patients with platinum-based chemotherapy in adjuvant (Olaussen et al, 2006; Zheng et al, 2007; Rosell et al, 2007; Simon et al, 2008; Bartolucci et al, 2009) or metastatic settings (Lord et al, 2002; Rosell et al, 2004; Ceppi et al, 2006; Wang et al, 2008; Rosell et al, 2009; Vilmar et al, 2010; Papadaki et al, 2011). Moreover, SNPs in DNA repair genes could represent predictive biomarkers as their presence could affect response to platinum-based therapy through different abilities to remove platinum-DNA adducts (Camps et al, 2007; Horgan et al, 2011). However, the great majority of data available are based on retrospective single-arm studies, not allowing to clearly dissect the prognostic vs the predictive role and, within this latter, to clarify whether the possible predictive role is truly specific for cisplatin or applicable to any kind of chemotherapy agent.
In the FAST trial, cisplatin-containing regimens demonstrated a statistically significant superiority in survival compared with platinum-free treatments (Boni et al, 2012). The availability of clinical outcome data of patients treated with or without cisplatin in a randomised trial allowed us to develop this retrospective sub-study, Bio-FAST, aiming to evaluate the clinical interaction between several biological factors and platinum- vs non-platinum-based chemotherapy.
The study of five different polymorphisms did not allow to identify a potential SNP with a predictive role. Only the genotype Gln/Gln of XRCC1 gene showed increased OS compared with genotype Arg/−, as well as a potential predictive role in PFS (test of interaction, P=0.029). The XRCC1 is a scaffold protein essential to the BER and single-strand breaks pathways. More than 60 SNPs have been identified in XRCC1 gene, of which the most extensively investigated is Arg399Gln (Liao et al, 2012). Several studies have shown that XRCC1 Arg399Gln was associated with a better treatment outcome in NSCLC patients who underwent platinum-based chemotherapy (Horgan et al, 2011), whereas no association was seen in non-platinum-treated patients. In a recent meta-analysis, XRCC1399Gln was partially favourably associated with both response rate and overall survival than 399Arg in analyses using all available studies; however, these associations became insignificant when only high-quality studies were considered (Wu et al, 2012).
Polymorphisms in DNA repair genes may contribute to interindividual diversity in DNA repair capacity; however, results from several studies have been generally inconsistent and obtained using low-quality genoty** methods. In 90 publications, the impact of genetic polymorphisms as predictive and/or prognostic markers in lung cancer has been modest at best (Horgan et al, 2011). A proportion of these studies, similar as ours, have been underpowered and the analysis of large number of polymorphisms raises the problem of multiple testing. There is also the possibility that inconsistent associations with SNPs may result from ethnic-related differences in allele frequencies. In fact, ERCC1 and XPD gene polymorphisms, recently, demonstrated better prognostic information for NSCLC patients, but in a large Chinese population (Zhang et al, 2012).
Patients were divided into ERCC1-negative and ERCC1-positive populations according to a cutoff value chosen a priori equal to that of the landmark study by Olaussen et al (2006). Regarding IHC expression of BRCA1, data available are more limited as almost all published studies evaluated BRCA1 mRNA levels (Taron et al, 2004; Rosell et al, 2007; Wang et al, 2008; Bartolucci et al, 2009; Rosell et al, 2009; Papadaki et al, 2011). Two studies determined IHC expression of BRCA1 in NSCLC and reported a higher percentage of positive tumours (90% and 84%, respectively) (Wachters et al, 2005; Ota et al, 2009). Moreover, in our study, there was a substantial discrepancy between ERCC1 and BRCA1 expression. This result is apparently in contrast with those in literature showing a tight correlation between mRNA levels of the two genes (Taron et al, 2004, Rosell et al, 2007; Wang et al, 2008; Bartolucci et al, 2009; Papadaki et al, 2012); however, these results are hardly comparable, considering a possible inconsistency between IHC protein and mRNA expression, as recently demonstrated (Friboulet et al, 2011). The disparities between IHC and mRNA can be explained not only by technical, but also biological reasons; in fact, considering the importance of post-transcriptional regulatory mechanisms (Friboulet et al, 2011), mRNA expression level could be very distant from the protein one.
As already reported, and in our study also, we found that ERCC1 (Olaussen et al, 2006; Vilmar et al, 2010) and BRCA1 (Rosell et al, 2007) levels were significantly higher in squamous cell carcinomas.
Our results regarding ERCC1 IHC expression allow us to confirm its strong prognostic role, considering that, overall, patients with ERCC1-negative tumours showed a significant advantage in response and survival than patients with ERCC1-positive tumours, as reported by other studies evaluating ERCC1 mRNA in advanced NSCLC patients treated with only platinum-based therapy (Lord et al, 2002; Rosell et al, 2004; Ceppi et al, 2006; Cobo et al, 2007; Hubner et al, 2011; Roth and Carlson, 2011). Concerning BRCA1, the positive patients showed a trend of better outcome than the negative ones. Our data are different from what was expected (Taron et al, 2004; Wang et al, 2008; Rosell et al, 2009; Papadaki et al, 2011; Papadaki et al, 2012), although it is necessary to emphasise that BRCA1 results could be influenced by the fact that it could also represent a predictive factor to vinorelbine (Quinn et al, 2003).
Considering the comparison between platinum and non-platinum therapy, unfortunately, we cannot conclude regarding the predictive role of either ERCC1 or BRCA1, because no statistically significant interaction between any clinical outcome and treatment delivered was found. Indeed, according to ERCC1 expression, a marginally statistically significant heterogeneity of the effect of treatment on response rate was observed. However, this event did not translate in any advantage in terms of duration of survival. Whether ERCC1 behaves as a simple generic prognostic factor or acts by predicting chemoresistance cannot be dissected from our study, given the lack of an untreated control arm.
The only study that allows to determine the prognostic vs predictive role of ERCC1 was carried out in the adjuvant setting (Olaussen et al, 2006). In this trial, high level of ERCC1 protein has been shown to be not only a positive prognostic factor in resected patients who did not receive adjuvant therapy, but also a negative predictive factor in patients treated with platinum chemotherapy. No similar study is available in advanced disease; moreover, it remains to be determined whether ERCC1 is only affecting cisplatin efficacy or that of any other drug. This would require a control arm with a non-platinum regimen as in our study.
A similar study assessed the predictive role of ERCC1 status for platinum-based chemotherapy in advanced NSCLC in the presence of an arm with non-platinum therapy (Rosell et al, 2004). A trend towards a better outcome in patients with low ERCC1 mRNA levels treated with cisplatin–gemcitabine, but not in other two arms, suggested that ERCC1 would be predictive of cisplatin–gemcitabine efficacy only. However, the robustness of this conclusion is compromised by retrospective design, small sample size and lack of a positive interaction test. The results of this study and of many others have led to the conclusion that ERCC1 expression could be used to select which patients should receive platinum-based chemotherapy, as prospective randomised trials are going to assess (Novello et al, 2012).
Our results, however, would not support this strategy, suggesting that ERCC1 expression could be used to identify which patients are going to derive most benefit from chemotherapy, regardless of the regimen administered. In line with this conclusion, Reynolds et al (2009) reported that ERCC1 expression, evaluated with RRM1, is predictive of response not only to carboplatin–gemcitabine therapy, but also to gemcitabine alone. It is therefore plausible that ERCC1 may also affect the efficacy of drugs different from platinum, such as gemcitabine. Indeed, it has to be noted that, in our trial, all patients received gemcitabine that might have affected the outcomes.
This study has some limitations. The number of unavailable tissue samples is relatively high, which is a common drawback in biomarker lung cancer studies. The main issue may arise from a selection bias. However, the cohorts of patients included and not included in the current evaluation did not significantly differ in clinical and biological characteristics at baseline and outcomes. In addition, statistical analyses adjusted for potential confounding factors showed results similar to those obtained in the unadjusted analyses. Moreover, patients were treated with four different regimens. Even if this could inflate the subgroup heterogeneity, FAST trial design allows to assess the predictive biomarker effect in the two platinum- vs the other two non-platinum-based regimens, and all statistical analyses, when appropriate, were performed including a covariate representing the absence/presence of the third drug. On the other side, it has to be emphasised that our study remains the only available study assessing the predictive role of ERCC1 IHC expression in a randomised trial with a non-cisplatin control arm.
In conclusion, our results confirm the prognostic role of ERCC1 expression and XRCC1 (Arg399Gln) polymorphism in advanced NSCLC treated with first-line chemotherapy. However, none of these biomarkers was shown to be a specific predictive factor of cisplatin efficacy. Further randomised controlled biomarker studies are needed to assess whether better tailoring of chemotherapy treatment may be accomplished with any novel tumour molecular marker.
Change history
30 April 2013
This paper was modified 12 months after initial publication to switch to Creative Commons licence terms, as noted at publication
References
Azzoli CG, Baker S, Temin S, Pao W, Aliff T, Brahmer J, Johnson DH, Laskin JL, Masters G, Milton D, Nordquist L, Pfister DG, Piantadosi S, Schiller JH, Smith R, Smith TJ, Strawn JR, Trent D, Giaccone G American Society of Clinical Oncology (2009) American Society of Clinical Oncology Clinical Practice Guideline update on chemotherapy for stage IV non-small-cell lung cancer. J Clin Oncol 27: 6251–6266.
Bartolucci R, Wei J, Sanchez JJ, Perez-Roca L, Chaib I, Puma F, Farabi R, Mendez P, Roila F, Okamoto T, Taron M, Rosell R (2009) XPG mRNA expression levels modulate prognosis in resected non-small-cell lung cancer in conjunction with BRCA1 and ERCC1 expression. Clin Lung Cancer 10: 47–52.
Boni C, Tiseo M, Boni L, Baldini E, Recchia F, Barone C, Grossi F, Germano D, Matano E, Marini G, Labianca R, Di Costanzo F, Bagnulo A, Pennucci C, Caroti C, Mencoboni M, Zanelli F, Prochilo T, Cafferata MA, Ardizzoni A Gruppo Oncologico Italiano di Ricerca Clinica (GOIRC) (2012) Triplets versus doublets, with or without cisplatin, in the first-line treatment of stage IIIB-IV non-small cell lung cancer (NSCLC) patients: a multicenter randomized factorial trial (FAST). Br J Cancer 106: 658–665.
Camps C, Domine M, Alberola V, Alonso G, Artal A, Gonzalez-Larriba JL, Gomez RG, Baron-Duarte FJ, Pujol-Obis E, Rosell R (2006) Clinical outcome of gemcitabine (GEM)/cisplatin (CIS) – vs docetaxel (DOC)/CIS – vs DOC/GEM treated stage IV Non-Small-Cell Lung Cancer (NSCLC) patients (P) according to X-Ray Repair Cross Complementing Group 3 (XRCC3) polymorphism and age. Ann Oncol 17 (Supplement9): ix213–ix240 doi:10.1093/annonc/mdl216.
Camps C, Sirera R, Iranzo V, Taron M, Rosell R (2007) Gene expression and polymorphisms of DNA repair enzymes: cancer susceptibility and response to chemotherapy. Clin Lung Cancer 8: 369–375.
Ceppi P, Volante M, Novello S, Rapa I, Danenberg KD, Danenberg PV, Cambieri A, Selvaggi G, Saviozzi S, Calogero R, Papotti M, Scagliotti GV (2006) ERCC1 and RRM1 gene expressions but not EGFR are predictive of shorter survival in advanced non-small-cell lung cancer treated with cisplatin and gemcitabine. Ann Oncol 17: 1818–1825.
Cobo M, Isla D, Massuti B, Montes A, Sanchez JM, Provencio M, Viñolas N, Paz-Ares L, Lopez-Vivanco G, Muñoz MA, Felip E, Alberola V, Camps C, Domine M, Sanchez JJ, Sanchez-Ronco M, Danenberg K, Taron M, Gandara D, Rosell R (2007) Customizing cisplatin based on quantitative excision repair cross-complementing 1 mRNA expression: a phase III trial in non-small-cell lung cancer. J Clin Oncol 25: 2747–2754.
de las Penas R, Sanchez-Ronco M, Alberola V, Taron M, Camps C, Garcia-Carbonero R, Massuti B, Queralt C, Botia M, Garcia-Gomez R, Isla D, Cobo M, Santarpia M, Cecere F, Mendez P, Sanchez JJ, Rosell R Spanish Lung Cancer Group (2006) Polymorphisms in DNA repair genes modulate survival in cisplatin/gemcitabine-treated non-small-cell lung cancer patients. Ann Oncol 17: 668–675.
Friboulet L, Barrios-Gonzales D, Commo F, Olaussen KA, Vagner S, Adam J, Goubar A, Dorvault N, Lazar V, Job B, Besse B, Validire P, Girard P, Lacroix L, Hasmats J, Dufour F, André F, Soria JC (2011) Molecular characteristics of ERCC1-negative versus ERCC1-positive tumors in resected NSCLC. Clin Cancer Res 17: 5562–5572.
Gazdar AF (2007) DNA repair and survival in lung cancer – the two faces of Janus. N Engl J Med 356: 771–773.
Gurubhagavatula S, Liu G, Park S, Zhou W, Su L, Wain JC, Lynch TJ, Neuberg DS, Christiani DC (2004) XPD and XRCC1 genetic polymorphisms are prognostic factors in advanced non–small-cell lung cancer patients treated with platinum chemotherapy. J Clin Oncol 22: 2594–2561.
Horgan AM, Yang B, Azad AK, Amir E, John T, Cescon DW, Wheatley-Price p, Hung RJ, Shepherd FA, Liu G (2011) Pharmacogenetic and germline prognostic markers of lung cancer. J Thoracic Oncol 6: 296–304.
Hubner RA, Riley RD, Billingham LJ, Popet S (2011) Excision repair cross-complementation group 1 (ERCC1) status and lung cancer outcomes: a meta-analysis of published studies and recommendations. PLoS One 6: e25164.
Husain A, He G, Venkatraman ES, Spriggs DR (1998) BRCA1 up-regulation is associated with repair-mediated resistance to cis-diamminedichloroplatinum (II). Cancer Res 58: 1120–1123.
Isla D, Sarries C, Rosell R, Alonso G, Domine M, Taron M, Lopez-Vivanco G, Camps C, Botia M, Nuñez L, Sanchez-Ronco M, Sanchez JJ, Lopez-Brea M, Barneto I, Paredes A, Medina B, Artal A, Lianes P (2004) Single nucleotide polymorphisms and outcome in docetaxel–cisplatin-treated advanced non-small-cell lung cancer. Ann Oncol 15: 1194–1203.
Joshi MB, Shirota Y, Danenberg KD, Conlon DH, Salonga DS, Herndon JE, Danenberg PV, Harpole DH (2005) High gene expression of TSI, GSTP1 and ERCC1 are risk factors for survival in patients treated with trimodality therapy for esophageal cancer. Clin Cancer Res 11: 2215–2221.
Kalikaki A, Kanaki M, Vassalou H, Souglakos J, Voutsina A, Georgoulias V, Mavroudis D (2009) DNA repair gene polymorphisms predict favorable clinical outcome in advanced non–small-cell lung cancer. Clin Lung Cancer 10: 118–123.
Li Q, Yu JJ, Mu C, Yunmbam MK, Slavsky D, Cross CL, Bostick-Bruton F, Reed E (2000) Association between the level of ERCC1 expression and the repair of cisplatin-induced DNA damage in human ovarian cancer cells. Anticancer Res 20: 645–652.
Liao WY, Shih JY, Chang GC, Cheng YK, Yang JC, Chen YM, Yu CJ (2012) Genetic polymorphism of XRCC1 Arg399Gln is associated with survival in non-small-cell lung cancer patients treated with gemcitabine/platinum. J Thorac Oncol 7: 973–981.
Lord RV, Brabender J, Gandara D, Alberola V, Camps C, Domine M, Cardenal F, Sánchez JM, Gumerlock PH, Tarón M, Sánchez JJ, Danenberg KD, Danenberg PV, Rosell R (2002) Low ERCC1 expression correlates with prolonged survival after cisplatin plus gemcitabine chemotherapy in non-small cell lung cancer. Clin Cancer Res 8: 2286–2291.
Martin LP, Hamilton TC, Schilder RJ (2008) Platinum resistance: the role of DNA repair pathways. Clin Cancer Res 14: 1291–1295.
Novello S, Manegold C, Grohe C, Colantonio I, Serke MH, Geissler M, Valmadre G, Schuette W, Stoelben E, Bria E, Monica V, Torri V, Scagliotti GV (2012) International tailored chemotherapy adjuvant trial: Itaca trial. J Clin Oncol 30, (suppl; abstract TPS7109).
Olaussen KA, Dunant A, Fouret P, Brambilla E, Andrè F, Haddad V, Taranchon E, Filipits M, Pirker R, Popper HH, Stahel R, Sabatier L, Pignon JP, Tursz T, Le Chevalier T, Soria JC IALT Bio Investigators (2006) DNA repair by ERCC1 in non-small-cell lung cancer and cisplatin-based adjuvant chemotherapy. N Engl J Med 355: 983–991.
Ota S, Ishii G, Goto K, Kubota K, Kim YH, Kojika M, Murata Y, Yamazaki M, Nishiwaki Y, Eguchi K, Ochiai A (2009) Immunohistochemical expression of BCRP and ERCC1 in biopsy specimen predicts survival in advanced non-small-cell lung cancer treated with cisplatin-based chemotherapy. Lung Cancer 64: 98–104.
Papadaki C, Tsaroucha E, Kaklamanis L, Lagoudaki E, Trypaki M, Tryfonidis K, Mavroudis D, Stathopoulos E, Georgoulias V, Souglakos J (2011) Correlation of BRCA1, TXR1 and TSP1 mRNA expression with treatment outcome to docetaxel-based first-line chemotherapy in patients with advanced/metastatic non-small-cell lung cancer. Br J Cancer 104: 316–323.
Papadaki C, Sfakianaki M, Ioannidis G, Lagoudaki E, Trypaki M, Tryfonidis K, Mavroudis D, Stathopoulos E, Georgoulias V, Souglakos J (2012) ERCC1 and BRAC1 mRNA expression levels in the primary tumor could predict the effectiveness of the second-line cisplatin-based chemotherapy in pretreated patients with metastatic non-small cell lung cancer. J Thoracic Oncol 7: 663–671.
Peters S, Adjei AA, Gridelli C, Reck M, Kerr K, Felip E ESMO Guidelines Working Group (2012) Metastatic non-small-cell lung cancer (NSCLC): ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann Oncol 23 (Suppl 7): vii56–vii64.
Quinn JE, Kennedy RD, Mullan PB, Gilmore PM, Carty M, Johnston PG, Harkin DP (2003) BRCA1 functions as a differential modulator of chemotherapy-induced apoptosis. Cancer Res 63: 6221–6228.
Reguart N, Cardona AF, Carrasco E, Gomez P, Taron M, Rosell R (2008) BRCA1: a new genomic marker for non-small-cell lung cancer. Clin Lung Cancer 9 (6): 331–339.
Reynolds C, Obasaju C, Schell MJ, Li X, Zheng Z, Boulware D, Caton JR, Demarco LC, O’Rourke MA, Shaw Wright G, Boehm KA, Asmar L, Bromund J, Peng G, Monberg MJ, Bepler G (2009) Randomized phase III trial of gemcitabine-based chemotherapy with in situ RRM1 and ERCC1 protein levels for response prediction in non-small-cell lung cancer. J Clin Oncol 27: 5808–5815.
Rosell R, Danenberg KD, Alberola V, Bepler G, Sanchez JJ, Camps C, Provencio M, Isla D, Taron M, Diz P, Artal A Spanish Lung Cancer Group (2004) Ribonucleotide reductase messenger RNA expression and survival in gemcitabine/cisplatin-treated advanced non-small cell lung cancer. Clin Cancer Res 10: 1318–1325.
Rosell R, Skrzypski M, Jassem E, Taron M, Bartolucci R, Sanchez JJ, Mendez P, Chaib I, Perez-Roca L, Szymanowska A, Rzyman W, Puma F, Kobierska-Gulida G, Farabi R, Jassem J (2007) BRCA1: a novel prognostic factor in resected non-small-cell lung cancer. PLoS One 2: e1129.
Rosell R, Perez-Roca L, Sanchez JJ, Cobo M, Moran T, Chaib I, Provencio M, Domine M, Sala MA, Jimenez U, Diz P, Barneto I, Macias JA, de Las Peñas R, Catot S, Isla D, Sanchez JM, Ibeas R, Lopez-Vivanco G, Oramas J, Mendez P, Reguart N, Blanco R, Taron M (2009) Customized treatment in non-small-cell lung cancer based on EGFR mutations and BRCA1 mRNA expression. PLoS One 4: e5133.
Roth JA, Carlson JJ (2011) Prognostic role of ERCC1 in advanced non-small-cell lung cancer: a systematic review and meta-analysis. Clin Lung Cancer 12: 393–401.
Ryu JS, Hong YC, Han HS, Lee JE, Kim S, Park YM, Kim YC, Hwang TS (2004) Association between polymorphisms of ERCC1 and XPD and survival in non-small-cell lung cancer patients treated with cisplatin combination chemotherapy. Lung Cancer 44: 311–316.
Simon GR, Sharma S, Cantor A, Smith P, Bepler G (2008) ERCC1 expression is a predictor of survival in resected patients with non-small cell lung cancer. Chest 127: 978–983.
Taron M, Rosell R, Felip E, Mendez P, Souglakos J, Ronco MS, Queralt C, Majo J, Sanchez JM, Sanchez JJ, Maestre J (2004) BRCA1 mRNA expression levels as an indicator of chemo-resistance in lung cancer. Hum Mol Genetic 13: 2443–2449.
Therasse P, Arbuck SG, Eisenhauer EA, Wanders J, Kaplan RS, Rubinstein L, Verweij J, Van Glabbeke M, van Oosterom AT, Christian MC, Gwyther SG (2000) New guidelines to evaluate the response to treatment in solid tumors: European Organization for Research and Treatment of Cancer, National Cancer Institute of the United States, National Cancer Institute of Canada. J Natl Cancer Inst 92: 205–216.
Vilmar AC, Santoni-Rugiu E, Sørensen JB (2010) ERCC1 and histopathology in advanced NSCLC patients randomized in a large multicenter phase III trial. Ann Oncol 21: 1817–1824.
Wachters FM, Wong LS, Timens W, Kam**a HH, Groen HJ (2005) ERCC1, hRad51 and BRCA1 protein expression in relation to tumour response and survival of stage III/IV NSCLC patients treated with chemotherapy. Lung Cancer 50: 211–219.
Wang L, Wei J, Qian X, Yin H, Zhao Y, Yu L, Wang T, Liu B (2008) ERCC1 and BRCA1 mRNA expression levels in metastatic malignant effusions is associated with chemosensitivity to cisplatin and/or docetaxel. BMC Cancer 8: 97.
Wei SZ, Zhan P, Shi MQ, Shi Y, Qian Q, Yu LK, Song Y (2011) Predictive value of ERCC1 and XPD polymorphism in patients with advanced non-small cell lung cancer receiving platinum based chemotherapy: a systematic review and meta-analysis. Med Oncol 28: 315–321.
Wu J, Liu J, Zhou Y, Ying J, Zou H, Guo S, Wang L, Zhao N, Hu J, Lu D, ** L, Li Q, Wang JC (2012) Predictive value of XRCC1 gene polymorphisms on platinum-based chemotherapy in advanced non-small cell lung cancer patients: a systematic review and meta-analysis. Clin Cancer Res 18: 3972–3981.
Yin M, Yan J, Voutsina A, Tibaldi C, Christiani DC, Heist RS, Rosell R, Botoon R, Wei Q (2011) No evidence of an association of ERCC1 and ERCC2 polymorphisms with clinical outcomes of platinum-based chemotherapies in non-small cell lung cancer: a meta-analysis. Lung Cancer 72: 370–377.
Zhang ZY, Tian X, Wu R, Liang Y, ** XY (2012) Predictive role of ERCC1 and XPD genetic polymorphisms in survival of Chinese non-small cell lung cancer patients receiving chemotherapy. Asian Pac J Cancer Prev 13: 2583–2586.
Zheng Z, Chen T, Li X, Haura E, Sharma A, Bepler G (2007) DNA synthesis and repair genes RRM1 and ERCC1 in lung cancer. N Engl J Med 356: 800–808.
Author information
Authors and Affiliations
Consortia
Corresponding author
Additional information
This work is published under the standard license to publish agreement. After 12 months the work will become freely available and the license terms will switch to a Creative Commons Attribution-NonCommercial-Share Alike 3.0 Unported License.
Supplementary Information accompanies this paper on British Journal of Cancer website
Supplementary information
Rights and permissions
This article is published under an open access license. Please check the 'Copyright Information' section either on this page or in the PDF for details of this license and what re-use is permitted. If your intended use exceeds what is permitted by the license or if you are unable to locate the licence and re-use information, please contact the Rights and Permissions team.
About this article
Cite this article
Tiseo, M., Bordi, P., Bortesi, B. et al. ERCC1/BRCA1 expression and gene polymorphisms as prognostic and predictive factors in advanced NSCLC treated with or without cisplatin. Br J Cancer 108, 1695–1703 (2013). https://doi.org/10.1038/bjc.2013.127
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1038/bjc.2013.127
- Springer Nature Limited