Abstract
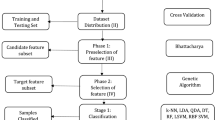
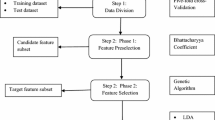
Correct classification of neuromuscular disorders is essential to provide accurate diagnosis. Presently, gene microarray technology is a widely accepted technology to monitor the expression level of a large number of genes simultaneously. The gene microarray data are a high dimensional data, which usually contains small samples having a large number of genes. Therefore, dimension reduction is a crucial task for correct classification of diseases. Dimension reduction eliminates those genes which are less expressive and enhances the efficiency of the classification model. In the present paper, we developed a novel hybrid dimension reduction method and a deep learning-based classification model for neuromuscular disorders. The hybrid dimension reduction method is deployed in three phase: in the first phase, the expressive genes are selected using F test method, and the mutual information method and the best one among them are selected for further processing. In second phase, the gene selected by the best model is further transformed to low dimension by PCA. In third phase, the deep learning-based classification model is deployed. For experimentation, two diseased and multi-diseased micro array data sets, which is publicly available, is used. The best accuracy by 50-100-50-25-13 deep learning architecture with hybrid dimension reduction, where 100 genes select by F test and PCA with 50 principal components is 89% for NMD data set. The best accuracy by 50-100-2 deep learning architecture with hybrid dimension reduction, where 100 genes select by F test and PCA with 50 principal components is 97% for FSHD data set. The proposed hybrid method gives better classification accuracy result and reduces the search space and time complexity as well for both two diseased and multi-diseased micro array data sets.

















Similar content being viewed by others
Data availability
The data sets analysed during the current study are gene expression profiles GSE36398 and E-GEOD-3307 obtained from NCBI website (https://www.ncbi.nlm.nih.gov).
References
Anand D, Pandey B, Pandey DK (2018) A novel hybrid feature selection model for classification of neuromuscular dystrophies using Bhattacharyya coefficient, genetic algorithm and radial basis function based support vector machine. Interdiscip Sci 10(2):244–250. https://doi.org/10.1007/s12539-016-0183-6 (Epub 2016 Sep 17)
Azuaje F (2000) Gene expression patterns and cancer classification: a self-adaptive and incremental neural approach. In: Proceedings of IEEE EMBS international conference on Information technology applications in biomedicine pp 308–313
Berrar DP, Downes CS, Dubitzky W (2002) Multiclass cancer classification using gene expression profiling and probabilistic neural networks. Biocomputing 8:5–16
Chen CK (2003) The classification of cancer stage microarray data. Comput Methods Programs Biomed 108(3):1070–1077
Gao L, Ye M, Lu X, Huang D (2017) Hybrid method based on information gain and support vector machine for gene selection in cancer classification. Genom Proteom Bioinform 15(6):389–395
Gonzalez-Navarro FF, Belanche-Munoz LA, Silva-Colon KA (2013) Effective classification and gene expression profiling for the facioscapulohumeral muscular dystrophy. PLoS ONE 8(12):e82071
Guyon I, Weston J, Barnhill S, Vapnik V (2002) Gene selection for cancer classification using support vector machines. Mach Learn 46(1–3):389–422
Hernandez JC, Duval B, Hao JK (2007) A genetic embedded approach for gene selection and classification of microarray data. In: Marchiori E, Moore JH, Rajapakse JC (eds) Evolutionary computation, machine learning data mining bioinformatics. 90–101.
Hira ZM, Gillies DF (2015) A review of feature selection and feature extraction methods applied on microarray data. AdvBioinform. https://doi.org/10.1155/2015/198363
Liu B, Cui Q, Jiang T, Ma S (2004) A combinational feature selection and ensemble neural network method for classification of gene expression data. BMC Bioinform 5(1):1
Lu H, Chen J, Yan K, ** Q, Xue Y, Gao Z (2017) A hybrid feature selection algorithm for gene expression data classification. Neurocomputing 256:56–62
Mohamad MS, Deris S, Yatim SM, Othman MR (2004) Feature selection method using genetic algorithm for the classification of small and high dimension data. In: Proceedings of the first international symposium on information and communication technology, 1–4:25.
Pashaei E, Pashaei E, Aydin N (2019) Gene selection using hybrid binary black hole algorithm and modified binary particle swarm optimization. Genomics 111(4):669–686
Peterson C, Ringner M (2003) Analyzing tumor gene expression profiles. ArtifIntell Med 28(1):59–74
Saeys Y, Inza I, Larranaga PA (2007) A review of feature selection techniques in bioinformatics. Bioinformatics 23(19):2507–2517
Schaefer G, Nakashima T (2010) Data mining of gene expression data by fuzzy and hybrid fuzzy methods. IEEE Trans InfTechnol Biomed 14(1):23–29
Sharbaf FV, Mosafer S, Moattar MH (2016) A hybrid gene selection approach for microarray data classification using cellular learning automata and ant colony optimization. Genomics 107(6):231–238
Sharma A, Paliwal KK (2008) Cancer classification by gradient LDA technique using microarray gene expression data. Data Knowl Eng 66(2):338–347
Shukla AK, Singh P, Vardhan M (2018) A hybrid gene selection method for microarray recognition. Biocybern Biomed Eng 38(4):975–991
Shukla AK, Singh P, Vardhan M (2020) Gene selection for cancer types classification using novel hybrid metaheuristics approach. Swarm Evol Comput 54:100661. https://doi.org/10.1016/j.swevo.2020.100661
Subasi A (2013) Classification of EMG signals using PSO optimized SVM for diagnosis of neuromuscular disorders. ComputBiol Med 43(5):576–586
Tang J, Alelyani S, Liu H (2014) Feature selection for classification: a review. Data Classif Algorithms Appl 37:1–29
Yao B, Li S (2010) ANMM4CBR: a case-based reasoning method for gene expression data classification. Algorithms Mol Biol 5(1):1
Zhang JG, Deng HW (2007) Gene selection for classification of microarray data based on the Bayes error. BMC Bioinform 8(1):370
Zheng CH, Chong YW, Wang HQ (2011) Gene selection using independent variable group analysis for tumor classification. Neural Comput Appl 20(2):161–170
Zibakhsh A, Abadeh MS (2013) Gene selection for cancer tumor detection using a novel memetic algorithm with a multi-view fitness function. Eng Appl Artif Intell 26(4):1274–1281
Acknowledgements
Dr. Babita Pandey is thankful to UGC-BSR start-up grant no F.30-460/2019 (BSR), India under which the current research work was carried out.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Pandey, B., Pandey, D.K., Khamparia, A. et al. A novel hybrid dimension reduction and deep learning-based classification for neuromuscular disorder. Adv. in Comp. Int. 2, 35 (2022). https://doi.org/10.1007/s43674-022-00047-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s43674-022-00047-7