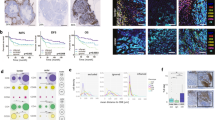
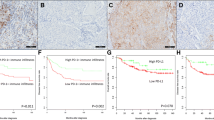
Abstract
Recently, immunotherapy has emerged as a promising and effective method for treating triple-negative breast cancer (TNBC). However, challenges still persist. Immunogenic cell death (ICD) is considered a prospective treatment and potential combinational treatment strategy as it induces an anti-tumor immune response by presenting the antigenic epitopes of dead cells. Nevertheless, the ICD process in TNBC and its impact on disease progression and the response to immunotherapy are not well understood. In this study, we observed dysregulation of the ICD process and verified the altered expression of prognostic ICD genes in TNBC through quantitative real-time polymerase chain reaction (qRT-PCR) analysis. To investigate the potential role of the ICD process in TNBC progression, we determined the ICD-dependent subtypes, and two were identified. Analysis of their distinct tumor immune microenvironment (TIME) and cancer hallmark features revealed that Cluster 1 and 2 corresponded to the immune “cold” and “hot” phenotypes, respectively. In addition, we constructed the prognostic signature ICD score of TNBC patients and demonstrated its clinical independence and generalizability. The ICD score could also serve as a potential biomarker for immune checkpoint blockade and may aid in the identification of targeted effective agents for individualized clinical strategies.





Similar content being viewed by others
Data Availability
The raw data supporting the conclusions of this article can be acquired from the public resources of TCGA (https://portal.gdc.cancer.gov) and the GEO (https://www.ncbi.nlm.nih.gov/geo/).
Abbreviations
- AUC:
-
Area under the receiver operating characteristic curve
- BC:
-
Breast cancer
- DEG:
-
Differentially expressed gene
- GEO:
-
Gene expression omnibus
- GO:
-
Gene ontology
- KEGG:
-
Kyoto encyclopedia of genes and genomes
- IC50:
-
Half-maximal inhibitory concentration
- ICD:
-
Immunogenic cell death
- OS:
-
Overall survival
- PCA:
-
Principal component analysis
- qRT-PCR:
-
Quantitative real-time polymerase chain reaction
- ROC:
-
Receiver operating characteristic
- ssGSEA:
-
Single-sample gene set enrichment analysis
- TCGA:
-
The Cancer Genome Atlas
- TIME:
-
Tumor immune microenvironment
- TNBC:
-
Triple-negative breast cancer
References
Abouelnazar FA, Zhang X, Wang M, Zhang J, Yu D, Zang X et al (2023) The new advance of SALL4 in cancer: function, regulation, and implication. J Clin Lab Anal. https://doi.org/10.1002/jcla.24927
Adams S, Gray RJ, Demaria S, Goldstein L, Perez EA, Shulman LN et al (2014) Prognostic value of tumor-infiltrating lymphocytes in triple-negative breast cancers from two phase III randomized adjuvant breast cancer trials: ECOG 2197 and ECOG 1199. J Clin Oncol 32(27):2959–2966. https://doi.org/10.1200/JCO.2013.55.0491
Asleh K, Brauer HA, Sullivan A, Lauttia S, Lindman H, Nielsen TO et al (2020) Predictive biomarkers for adjuvant capecitabine benefit in early-stage triple-negative breast cancer in the FinXX Clinical Trial. Clin Cancer Res 26(11):2603–2614. https://doi.org/10.1158/1078-0432.CCR-19-1945
Cheang MC, Voduc D, Bajdik C, Leung S, McKinney S, Chia SK et al (2008) Basal-like breast cancer defined by five biomarkers has superior prognostic value than triple-negative phenotype. Clin Cancer Res 14(5):1368–1376. https://doi.org/10.1158/1078-0432.CCR-07-1658
Diana A, Carlino F, Franzese E, Oikonomidou O, Criscitiello C, De Vita F et al (2020) Early triple negative breast cancer: conventional treatment and emerging therapeutic landscapes. Cancers (basel). https://doi.org/10.3390/cancers12040819
Foulkes WD, Smith IE, Reis-Filho JS (2010) Triple-negative breast cancer. N Engl J Med 363(20):1938–1948. https://doi.org/10.1056/NEJMra1001389
Galluzzi L, Buque A, Kepp O, Zitvogel L, Kroemer G (2017) Immunogenic cell death in cancer and infectious disease. Nat Rev Immunol 17(2):97–111. https://doi.org/10.1038/nri.2016.107
Garg AD, Dudek-Peric AM, Romano E, Agostinis P (2015) Immunogenic cell death. Int J Dev Biol 59(1–3):131–140. https://doi.org/10.1387/ijdb.150061pa
Garg AD, De Ruysscher D, Agostinis P (2016) Immunological metagene signatures derived from immunogenic cancer cell death associate with improved survival of patients with lung, breast or ovarian malignancies: a large-scale meta-analysis. Oncoimmunology 5(2):e1069938. https://doi.org/10.1080/2162402X.2015.1069938
Garg AD, More S, Rufo N, Mece O, Sassano ML, Agostinis P et al (2017) Trial watch: immunogenic cell death induction by anticancer chemotherapeutics. Oncoimmunology 6(12):e1386829. https://doi.org/10.1080/2162402X.2017.1386829
Hudis CA, Gianni L (2011) Triple-negative breast cancer: an unmet medical need. Oncologist 16(1):1–11. https://doi.org/10.1634/theoncologist.2011-S1-01
Keenan TE, Tolaney SM (2020) Role of immunotherapy in triple-negative breast cancer. J Natl Compr Canc Netw 18(4):479–489. https://doi.org/10.6004/jnccn.2020.7554
Kroemer G, Galluzzi L, Kepp O, Zitvogel L (2013) Immunogenic cell death in cancer therapy. Annu Rev Immunol 31:51–72. https://doi.org/10.1146/annurev-immunol-032712-100008
Kroemer G, Galassi C, Zitvogel L, Galluzzi L (2022) Immunogenic cell stress and death. Nat Immunol 23(4):487–500. https://doi.org/10.1038/s41590-022-01132-2
Krysko DV, Garg AD, Kaczmarek A, Krysko O, Agostinis P, Vandenabeele P (2012) Immunogenic cell death and DAMPs in cancer therapy. Nat Rev Cancer 12(12):860–875. https://doi.org/10.1038/nrc3380
Lyons TG (2019) Targeted Therapies for Triple-Negative Breast Cancer. Curr Treat Options Oncol 20(11):82. https://doi.org/10.1007/s11864-019-0682-x
O’Reilly D, Sendi MA, Kelly CM (2021) Overview of recent advances in metastatic triple negative breast cancer. World J Clin Oncol 12(3):164–182. https://doi.org/10.5306/wjco.v12.i3.164
Peng Y, Shui L, **e J, Liu S (2020) Development and validation of a novel 15-CpG-based signature for predicting prognosis in triple-negative breast cancer. J Cell Mol Med 24(16):9378–9387. https://doi.org/10.1111/jcmm.15588
Pfirschke C, Engblom C, Rickelt S, Cortez-Retamozo V, Garris C, Pucci F et al (2016) Immunogenic chemotherapy sensitizes tumors to checkpoint blockade therapy. Immunity 44(2):343–354. https://doi.org/10.1016/j.immuni.2015.11.024
Spisek R, Dhodapkar MV (2007) Towards a better way to die with chemotherapy: role of heat shock protein exposure on dying tumor cells. Cell Cycle 6(16):1962–1965. https://doi.org/10.4161/cc.6.16.4601
Telli ML, Timms KM, Reid J, Hennessy B, Mills GB, Jensen KC et al (2016) Homologous recombination deficiency (HRD) Score predicts response to platinum-containing neoadjuvant chemotherapy in patients with triple-negative breast cancer. Clin Cancer Res 22(15):3764–3773. https://doi.org/10.1158/1078-0432.CCR-15-2477
Wang X, Wu S, Liu F, Ke D, Wang X, Pan D et al (2021) An immunogenic cell death-related classification predicts prognosis and response to immunotherapy in head and neck squamous cell carcinoma. Front Immunol 12:781466. https://doi.org/10.3389/fimmu.2021.781466
Wei LY, Zhang XJ, Wang L, Hu LN, Zhang XD, Li L et al (2020) A six-epithelial-mesenchymal transition gene signature may predict metastasis of triple-negative breast cancer. Onco Targets Ther 13:6497–6509. https://doi.org/10.2147/OTT.S256818
Wilson TR, Yu J, Lu X, Spoerke JM, **ao Y, O’Brien C et al (2016) The molecular landscape of high-risk early breast cancer: comprehensive biomarker analysis of a phase III adjuvant population. NPJ Breast Cancer 2:16022. https://doi.org/10.1038/npjbcancer.2016.22
Yin L, Duan JJ, Bian XW, Yu SC (2020) Triple-negative breast cancer molecular subty** and treatment progress. Breast Cancer Res 22(1):61. https://doi.org/10.1186/s13058-020-01296-5
Zitvogel L, Galluzzi L, Smyth MJ, Kroemer G (2013) Mechanism of action of conventional and targeted anticancer therapies: reinstating immunosurveillance. Immunity 39(1):74–88. https://doi.org/10.1016/j.immuni.2013.06.014
Funding
This work was supported by the National Natural Science Foundation of China (82205114) and the Natural Science Foundation of Shandong Province (ZR2020MH356).
Author information
Authors and Affiliations
Contributions
SL, XFL, YNQ, and CYW conceived and designed the experiments. YYS, YYW, FFL, and KXJ analyzed the data, prepared figures and tables. XFF, YW, XYS, RW, LXC and JZZ prepared figures and/or tables. SL and YYS authored or reviewed drafts of the paper. All authors approved the final version of the manuscript.
Corresponding authors
Ethics declarations
Conflict of interest
No potential conflict of interest was reported by the author(s).
Ethical Approval
The studies involving human participants were reviewed and approved by the ethics committee of the Longhua Hospital Affiliated to Shanghai University of Traditional Chinese Medicine (NO: 2021-043). The patients/participants provided their written informed consent to participate in this study.
Consent to Participate
All the participants provided written informed consent.
Consent for Publish
The participants consent to have their data published.
Supplementary Information
Below is the link to the electronic supplementary material.
43657_2023_133_MOESM1_ESM.docx
Supplementary file1 Fig. S1 The co-expression analysis of ICD genes in TNBC. a Results of the TCGA TNBC samples. b Results of the TNBC cohorts (GSE103091). Fig. S2 The prognostic value of ICD genes in TNBC. a Univariate Cox proportional hazards regression analysis results of all 34 ICD genes. b The K-M curves of the OS according to the expression group of ICD genes, where the median expression was the cut-off. c The K-M curves of MFS according to the expression group of ICD genes, where the median expression was the cut-off. Fig. S3 The differential expression of ICD-related genes in malignant versus non-malignant epithelial cells. a Harmony removes sample batches: This component highlights the use of the Harmony algorithm to mitigate batch effects between samples. b Cell clustering and annotation results (including subsets 0, 2, 4, 6, and 7 as epithelial cells): This component presents the results of cell clustering and manual annotation, specifically identifying epithelial cell subsets. c Box plot of differential expression of ICD-related genes: This component utilizes a box plot to visualize the differential expression of ICD-related genes between malignant and non-malignant epithelial cells. d Heat map of differential expression of ICD-related genes: This component employs a heat map to illustrate the patterns of differential expression among the ICD-related genes in malignant and non-malignant epithelial cells (DOCX 3067 KB)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Shi, Y., Wu, Y., Li, F. et al. Investigating the Immunogenic Cell Death-Dependent Subtypes and Prognostic Signature of Triple-Negative Breast Cancer. Phenomics 4, 34–45 (2024). https://doi.org/10.1007/s43657-023-00133-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43657-023-00133-x