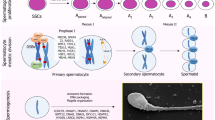
Abstract
Purpose of Review
Male factor infertility is common and often multifactorial. A subset of these patients have underlying genetic etiologies. CRISPR/Cas9 is a simple and flexible gene editing tool with promising applications in this space. This review aims to summarize the advances that CRISPR/Cas9-based tools have brought to the field and propose future directions for study.
Recent Findings
CRISPR/Cas9 has been applied successfully to spermatogonial stem cells (SSCs) and via pronuclear injection of zygotes to generate animal models of male factor infertility. These approaches have led to the high-throughput validation of candidate male fertility genes obtained either through genome-wide associated studies or testis-specific gene expression studies. One group has applied this further to SSCs in the correction of genetic infertility due to a mutation in the Kit gene.
Summary
Application of CRISPR/Cas9 to the investigation and treatment of male infertility holds promise in identifying novel genetic causes of NOA. Gene editing in germ cells to treat genetic infertility is technically feasible, but has not been used in humans due to significant ethical concerns. Stringent regulations are imperative to ensure safe translation of this technology into human populations.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: • Of importance •• Of major importance
Charo RA, Hynes RO, Beier DW, Clayton EW, Coller BS, Evans JH, et al. Human Genome Editing. 2017. Washington, DC: The National Academies Press.
Barrangou R, Fremaux C, Deveau H, Richards M, Boyaval P, Moineau S, et al. CRISPR provides acquired resistance against viruses in prokaryotes. Science (New York, NY). 2007 Mar 23;315(5819):1709–12.
Deltcheva E, Chylinski K, Sharma CM, Gonzales K, Chao Y, Pirzada ZA, et al. CRISPR RNA maturation by trans-encoded small RNA and host factor RNase III. Nature. 2011 Mar 31;471(7340):602–7.
**ek M, Jiang F, Taylor DW, Sternberg SH, Kaya E, Ma E, et al. Structures of Cas9 endonucleases reveal RNA-mediated conformational activation. Science (New York, NY) [Internet]. 2014 Mar 14;343(6176):1247997–1247997. Available from: http://www.sciencemag.org/cgi/doi/10.1126/science.1247997
Mohanraju P, Makarova KS, Zetsche B, Zhang F, Koonin EV, Oost J van der. Diverse evolutionary roots and mechanistic variations of the CRISPR-Cas systems. Science (New York, NY). 2016 Aug 5;353(6299):aad5147.
**ek M, Chylinski K, Fonfara I, Hauer M, Doudna JA, Charpentier E. A programmable dual-RNA-guided DNA endonuclease in adaptive bacterial immunity. Science (New York, NY) [Internet]. 2012 Aug 17;337(6096):816–21. Available from: http://www.sciencemag.org/cgi/doi/10.1126/science.1225829
Mojica FJM, Díez-Villaseñor C, García-Martínez J, Almendros C. Short motif sequences determine the targets of the prokaryotic CRISPR defence system. Microbiology (Reading, England). 2009 Mar;155(Pt 3):733–40.
Chen F, Pruett-Miller SM, Davis GD. Methods in molecular biology. Methods Mol Biology Clifton N J. 2014;1239:251–65.
Kim YG, Cha J, Chandrasegaran S. Hybrid restriction enzymes: zinc finger fusions to Fok I cleavage domain. Proc National Acad Sci. 1996;93(3):1156–60.
Wolfe SA, Nekludova L, Pabo CO. DNA recognition by Cys 2 His 2 zinc finger proteins. Annu Rev Bioph Biom. 2000;29(1):183–212.
Bitinaite J, Wah DA, Aggarwal AK, Schildkraut I. FokI dimerization is required for DNA cleavage. Proc National Acad Sci. 1998;95(18):10570–5.
Miller JC, Tan S, Qiao G, Barlow KA, Wang J, **a DF, et al. A TALE nuclease architecture for efficient genome editing. Nat Biotechnol. 2010;29(2):143–8.
Deng D, Yan C, Pan X, Mahfouz M, Wang J, Zhu J-K, et al. Structural basis for sequence-specific recognition of DNA by TAL effectors. Science. 2012;335(6069):720–3.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science (New York, NY). 2013 Feb 15;339(6121):819–23.
Mali P, Yang L, Esvelt KM, Aach J, Guell M, DiCarlo JE, et al. RNA-guided human genome engineering via Cas9. Science (New York, NY). 2013 Feb 15;339(6121):823–6.
Shalem O, Sanjana NE, Hartenian E, Shi X, Scott DA, Mikkelsen TS, et al. Genome-scale CRISPR-Cas9 knockout screening in human cells. Science (New York, NY). 2014 Jan 3;343(6166):84–7.
Wang T, Wei JJ, Sabatini DM, Lander ES. Genetic screens in human cells using the CRISPR-Cas9 system. Science (New York, NY). 2014 Jan 3;343(6166):80–4.
Cho SW, Kim S, Kim JM, Kim J-S. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nature biotechnology. 2013 Mar;31(3):230–2.
Bikard D, Jiang W, Samai P, Hochschild A, Zhang F, Marraffini LA. Programmable repression and activation of bacterial gene expression using an engineered CRISPR-Cas system. Nucleic acids research. 2013 Aug;41(15):7429–37.
Qi LS, Larson MH, Gilbert LA, Doudna JA, Weissman JS, Arkin AP, et al. Repurposing CRISPR as an RNA-guided platform for sequence-specific control of gene expression. Cell. 2013 Feb 28;152(5):1173–83.
Maeder ML, Linder SJ, Cascio VM, Fu Y, Ho QH, Joung JK. CRISPR RNA-guided activation of endogenous human genes. Nature methods. 2013 Oct;10(10):977–9.
Perez-Pinera P, Kocak DD, Vockley CM, Adler AF, Kabadi AM, Polstein LR, et al. RNA-guided gene activation by CRISPR-Cas9-based transcription factors. Nature methods. 2013 Oct;10(10):973–6.
Sander JD, Joung JK. CRISPR-Cas systems for editing, regulating and targeting genomes. Nature biotechnology [Internet]. 2014 Apr 1;32(4):347–55. Available from: https://www.nature.com/articles/nbt.2842
Liu XS, Wu H, Ji X, Stelzer Y, Wu X, Czauderna S, et al. Editing DNA Methylation in the mammalian genome. Cell. 2016 Sep 22;167(1):233–247.e17.
Hilton IB, D’Ippolito AM, Vockley CM, Thakore PI, Crawford GE, Reddy TE, et al. Epigenome editing by a CRISPR-Cas9-based acetyltransferase activates genes from promoters and enhancers. Nature biotechnology. 2015 May;33(5):510–7.
Pattanayak V, Lin S, Guilinger JP, Ma E, Doudna JA, Liu DR. High-throughput profiling of off-target DNA cleavage reveals RNA-programmed Cas9 nuclease specificity. Nature biotechnology [Internet]. 2013 Sep 1;31(9):839–43. Available from: http://www.nature.com/doifinder/10.1038/nbt.2673
Charlesworth CT, Deshpande PS, Dever DP, Camarena J, Lemgart VT, Cromer MK, et al. Identification of preexisting adaptive immunity to Cas9 proteins in humans. Nat Med. 2019;25(2):249–54.
Wagner DL, Amini L, Wendering DJ, Burkhardt L-M, Akyüz L, Reinke P, et al. High prevalence of Streptococcus pyogenes Cas9-reactive T cells within the adult human population. Nat Med. 2019;25(2):242–8.
Kumar N, Singh AK. Trends of male factor infertility, an important cause of infertility: a review of literature. J Hum Reproductive Sci. 2015;8(4):191–6.
Baker K, Jr. ES. Obstructive azoospermia: reconstructive techniques and results. Clinics. 2013;68(Suppl 1):61–73.
Chiba K, Enatsu N, Fujisawa M. Management of non-obstructive azoospermia. Reproductive Medicine Biology. 2016;15(3):165–73.
Esteves S, Agarwal A. Reproductive outcomes, including neonatal data, following sperm injection in men with obstructive and nonobstructive azoospermia: case series and systematic review. Clinics. 2013;68(S1):141–9.
Bhagavath B, Podolsky RH, Ozata M, Bolu E, Bick DP, Kulharya A, et al. Clinical and molecular characterization of a large sample of patients with hypogonadotropic hypogonadism. Fertil Steril. 2006;85(3):706–13.
Lanfranco F, Kamischke A, Zitzmann M, Nieschlag E. Klinefelter’s syndrome. Lancet. 2004;364(9430):273–83.
SCHWEIKERT HU, WEISSBACH L, LEYENDECKER G, SCHWINGER E, WARTENBERG H, KRÜCK F. Clinical, endocrinological, and cytological characterization of Two 46,XX Males*. J Clin Endocrinol Metabolism. 1982;54(4):745–52.
Stouffs K, Tournaye H, Liebaers I, Lissens W. Male infertility and the involvement of the X chromosome. Hum Reprod Update. 2009;15(6):623–37.
Telvi L, Lebbar A, Pino OD, Barbet JP, Chaussain JL. 45,X/46,XY Mosaicism: Report of 27 Cases. Pediatrics. 1999;104(2):304–8.
Colaco S, Modi D. Genetics of the human Y chromosome and its association with male infertility. Reprod Biol Endocrin. 2018;16(1):14.
Ghieh F, Mitchell V, Mandon-Pepin B, Vialard F. Genetic defects in human azoospermia. Basic Clin Androl. 2019;29(1):4.
Vij SC, Jr ES, Agarwal A. Biological therapy for non-obstructive azoospermia. Expert Opin Biol Th. 2017;18(1):1–5.
Sadri-Ardekani H, Atala A. Testicular tissue cryopreservation and spermatogonial stem cell transplantation to restore fertility: from bench to bedside. Stem Cell Res Ther. 2014;5(3):68.
Brinster RL, Zimmermann JW. Spermatogenesis following male germ-cell transplantation. Proc National Acad Sci. 1994;91(24):11298–302.
Nóbrega RH, Greebe CD, Kant H van de, Bogerd J, França LR de, Schulz RW. Spermatogonial stem cell niche and spermatogonial stem cell transplantation in zebrafish. Plos One. 2010;5(9):e12808.
Honaramooz A, Megee SO, Dobrinski I. Germ cell transplantation in pigs1. Biol Reprod. 2002;66(1):21–8.
Izadyar F, Ouden KD, Stout T, Stout J, Coret J, Lankveld D, et al. Autologous and homologous transplantation of bovine spermatogonial stem cells. Reproduction. 2003;126(6):765–74.
Schlatt S, Rosiepen G, Weinbauer GF, Rolf C, Brook PF, Nieschlag E. Germ cell transfer into rat, bovine, monkey and human testes. Hum Reprod. 1999;14(1):144–50.
Hermann BP, Sukhwani M, Winkler F, Pascarella JN, Peters KA, Sheng Y, et al. Spermatogonial stem cell transplantation into rhesus testes regenerates spermatogenesis producing functional sperm. Cell Stem Cell. 2012;11(5):715–26.
Honaramooz A, Snedaker A, Boiani M, Schöler H, Dobrinski I, Schlatt S. Sperm from neonatal mammalian testes grafted in mice. Nature. 2002;418(6899):778–81.
Yuan Z, Hou R, Wu J. Generation of mice by transplantation of an adult spermatogonial cell line after cryopreservation. Cell Proliferat. 2009;42(2):123–31.
Kubota H, Avarbock MR, Schmidt JA, Brinster RL. Spermatogonial stem cells derived from infertile Wv/Wv mice self-renew in vitro and generate progeny following transplantation. Biol Reprod. 2009;81(2):293–301.
Goossens E, Vos P de, Tournaye H. Array comparative genomic hybridization analysis does not show genetic alterations in spermatozoa and offspring generated after spermatogonial stem cell transplantation in the mouse. Hum Reprod. 2010;25(7):1836–42.
Wu X, Goodyear SM, Abramowitz LK, Bartolomei MS, Tobias JW, Avarbock MR, et al. Fertile offspring derived from mouse spermatogonial stem cells cryopreserved for more than 14 years. Hum Reprod. 2012;27(5):1249–59.
Honaramooz A, Behboodi E, Megee SO, Overton SA, Galantino-Homer H, Echelard Y, et al. Fertility and germline transmission of donor haplotype following germ cell transplantation in immunocompetent goats1. Biol Reprod. 2003;69(4):1260–4.
Herrid M, Olejnik J, Jackson M, Suchowerska N, Stockwell S, Davey R, et al. Irradiation enhances the efficiency of testicular germ cell transplantation in sheep. Biol Reprod. 2009;81(5):898–905.
Wu Y, Zhou H, Fan X, Zhang Y, Zhang M, Wang Y, et al. Correction of a genetic disease by CRISPR-Cas9-mediated gene editing in mouse spermatogonial stem cells. Cell Res. 2015;25(1):67–79.
Sato T, Sakuma T, Yokonishi T, Katagiri K, Kamimura S, Ogonuki N, et al. Genome editing in mouse spermatogonial stem cell lines using TALEN and double-nicking CRISPR/Cas9. Stem Cell Rep. 2015;5(1):75–82.
Chapman KM, Medrano GA, Jaichander P, Chaudhary J, Waits AE, Nobrega MA, et al. Targeted germline modifications in rats using CRISPR/Cas9 and spermatogonial stem cells. Cell Reports. 2015;10(11):1828–35.
Chen J, Cai T, Zheng C, Lin X, Wang G, Liao S, et al. MicroRNA-202 maintains spermatogonial stem cells by inhibiting cell cycle regulators and RNA binding proteins. Nucleic Acids Res. 2016;45(7):gkw1287.
Huang E, Nocka K, Beier DR, Chu T-Y, Buck J, Lahm H-W, et al. The hematopoietic growth factor KL is encoded by the SI locus and is the ligand of the c-kit receptor, the gene product of the W locus. Cell. 1990;63(1):225–33.
Li X, Sun T, Wang X, Tang J, Liu Y. Restore natural fertility of Kitw/Kitwv mouse with nonobstructive azoospermia through gene editing on SSCs mediated by CRISPR-Cas9. Stem Cell Res Ther. 2019;10(1):271.
Lim JJ, Sung S -Y., Kim HJ, Song S -H., Hong JY, Yoon TK, et al. Long-term proliferation and characterization of human spermatogonial stem cells obtained from obstructive and non-obstructive azoospermia under exogenous feeder-free culture conditions. Cell Proliferat. 2010;43(4):405–17.
Kosova G, Scott NM, Niederberger C, Prins GS, Ober C. Genome-wide association study identifies candidate genes for male fertility traits in humans. Am J Hum Genetics. 2012;90(6):950–61.
Aston KI, Carrell DT. Genome-wide study of single-nucleotide polymorphisms associated with azoospermia and severe oligozoospermia. J Androl. 2009;30(6):711–25.
Yu J, Wu H, Wen Y, Liu Y, Zhou T, Ni B, et al. Identification of seven genes essential for male fertility through a genome-wide association study of non-obstructive azoospermia and RNA interference-mediated large-scale functional screening in Drosophila. Hum Mol Genet. 2015;24(5):1493–503.
Aston KI, Krausz C, Laface I, Ruiz-Castané E, Carrell DT. Evaluation of 172 candidate polymorphisms for association with oligozoospermia or azoospermia in a large cohort of men of European descent. Hum Reprod. 2010;25(6):1383–97.
Parada-Bustamante A, Lardone MC, Valdevenito R, Ebensperger M, López PV, Madariaga M, et al. Analysis of 6 single-nucleotide polymorphisms in the androgen receptor gene in chilean patients with primary spermatogenic failure. J Androl. 2012;33(1):88–95.
He X-J, Ruan J, Du W-D, Chen G, Zhou Y, Xu S, et al. PRM1 variant rs35576928 (Arg>Ser) is associated with defective spermatogenesis in the Chinese Han population. Reprod Biomed Online. 2012;25(6):627–34.
Coutton C, Escoffier J, Martinez G, Arnoult C, Ray PF. Teratozoospermia: spotlight on the main genetic actors in the human. Hum Reprod Update. 2015;21(4):455–85.
Young SAM, Miyata H, Satouh Y, Kato H, Nozawa K, Isotani A, et al. CRISPR/Cas9-mediated rapid generation of multiple mouse lines identified Ccdc63 as essential for spermiogenesis. Int J Mol Sci. 2015;16(10):24732–50.
Maruyama S, Ito M, Ikami Y, Okitsu Y, Ito C, Toshimori K, et al. A critical role of solute carrier 22a14 in sperm motility and male fertility in mice. Sci Rep-uk. 2016;6(1):36468.
Young SAM, Miyata H, Satouh Y, Aitken RJ, Baker MA, Ikawa M. CABYR is essential for fibrous sheath integrity and progressive motility in mouse spermatozoa. J Cell Sci. 2016;129(23):4379–87.
Tang S, Wang X, Li W, Yang X, Li Z, Liu W, et al. Biallelic Mutations in CFAP43 and CFAP44 cause male infertility with multiple morphological abnormalities of the sperm flagella. Am J Hum Genetics. 2017;100(6):854–64.
Imtiaz A, Belyantseva IA, Beirl AJ, Fenollar-Ferrer C, Bashir R, Bukhari I, et al. CDC14A phosphatase is essential for hearing and male fertility in mouse and human. Hum Mol Genet. 2017;27(5):780–98.
Abbasi F, Miyata H, Shimada K, Morohoshi A, Nozawa K, Matsumura T, et al. Radial spoke head 6 homolog A is required for sperm flagellum formation and male fertility in mice. J Cell Sci. 2018;131(19):jcs.221648.
Kruger AN, Brogley MA, Huizinga JL, Kidd JM, Rooij DG de, Hu Y-C, et al. A Neofunctionalized X-linked ampliconic gene family is essential for male fertility and equal sex ratio in mice. Curr Biol. 2019;
Coutton C, Martinez G, Kherraf Z-E, Amiri-Yekta A, Boguenet M, Saut A, et al. Bi-allelic mutations in ARMC2 lead to severe astheno-teratozoospermia due to sperm flagellum malformations in humans and mice. Am J Hum Genetics. 2019;104(2):331–40.
Liu W, He X, Yang S, Zouari R, Wang J, Wu H, et al. Bi-allelic mutations in TTC21A induce asthenoteratospermia in humans and mice. Am J Hum Genetics. 2019;104(Hum. Reprod. Update 21 2015):738–48.
Li W, Wu H, Li F, Tian S, Kherraf Z-E, Zhang J, et al. Biallelic mutations in CFAP65 cause male infertility with multiple morphological abnormalities of the sperm flagella in humans and mice. J Med Genet. 2019;jmedgenet-2019-106344.
Shen Y, Zhang F, Li F, Jiang X, Yang Y, Li X, et al. Loss-of-function mutations in QRICH2 cause male infertility with multiple morphological abnormalities of the sperm flagella. Nat Commun. 2019;10(1):433.
He X, Li W, Wu H, Lv M, Liu W, Liu C, et al. Novel homozygous CFAP69 mutations in humans and mice cause severe asthenoteratospermia with multiple morphological abnormalities of the sperm flagella. J Med Genet. 2019;56(2):96.
Miyata H, Castaneda JM, Fujihara Y, Yu Z, Archambeault DR, Isotani A, et al. Genome engineering uncovers 54 evolutionarily conserved and testis-enriched genes that are not required for male fertility in mice. Proc National Acad Sci. 2016;113(28):7704–10.
Singh P, Schimenti JC. The genetics of human infertility by functional interrogation of SNPs in mice. Proc National Acad Sci. 2015;112(33):10431–6.
Sakib S, Goldsmith T, Voigt A, Dobrinski I. Testicular organoids to study cell–cell interactions in the mammalian testis. Andrology-us. 2019;
Zhou Y, Zhu S, Cai C, Yuan P, Li C, Huang Y, et al. High-throughput screening of a CRISPR/Cas9 library for functional genomics in human cells. Nature [Internet]. 2014 May 22;509(7501):487–91. Available from: q.
Hart T, Chandrashekhar M, Aregger M, Steinhart Z, Brown KR, MacLeod G, et al. High-resolution CRISPR screens reveal fitness genes and genotype-specific cancer liabilities. Cell. 2015;163(6):1515–26.
Mair B, Tomic J, Masud SN, Tonge P, Weiss A, Usaj M, et al. Essential gene profiles for human pluripotent stem cells identify uncharacterized genes and substrate dependencies. Cell Reports. 2019;27(2):599–615.e12.
Konermann S, Brigham MD, Trevino AE, Joung J, Abudayyeh OO, Barcena C, et al. Genome-scale transcriptional activation by an engineered CRISPR-Cas9 complex. Nature. 2015;517(7536):583–8.
Gilbert LA, Horlbeck MA, Adamson B, Villalta JE, Chen Y, Whitehead EH, et al. Genome-scale CRISPR-mediated control of gene repression and activation. Cell. 2014 Oct 23;159(3):647–61.
Klann TS, Black JB, Chellappan M, Safi A, Song L, Hilton IB, et al. CRISPR–Cas9 epigenome editing enables high-throughput screening for functional regulatory elements in the human genome. Nat Biotechnol. 2017;35(6):561–8.
Dong MB, Wang G, Chow RD, Ye L, Zhu L, Dai X, et al. Systematic immunotherapy target discovery using genome-scale in vivo CRISPR screens in CD8 T cells. Cell. 2019;178(5):1189–1204.e23.
Patel SJ, Sanjana NE, Kishton RJ, Eidizadeh A, Vodnala SK, Cam M, et al. Identification of essential genes for cancer immunotherapy. Nature. 2017;548(7669):537–42.
Manguso RT, Pope HW, Zimmer MD, Brown FD, Yates KB, Miller BC, et al. In vivo CRISPR screening identifies Ptpn2 as a cancer immunotherapy target. Nature. 2017 Jul 27;547(7664):413–8.
Jaitin DA, Weiner A, Yofe I, Lara-Astiaso D, Keren-Shaul H, David E, et al. Dissecting immune circuits by linking CRISPR-pooled screens with single-cell RNA-Seq. Cell. 2016;167(7):1883–1896.e15.
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell [Internet]. 2013 May;153(4):910–8. Available from: http://www.sciencedirect.com/science/article/pii/S0092867413004674
Liang P, Xu Y, Zhang X, Ding C, Huang R, Zhang Z, et al. CRISPR/Cas9-mediated gene editing in human tripronuclear zygotes. Protein Cell. 2015;6(5):363–72.
Charo A, Hynes RO, Beier DW, Clayton EW, Coller BS, Evans JH, et al. Human genome editing: science, ethics, and governance. The National Academies of Sciences, Engineering, and Medicine; 2017.
Ma H, Marti-Gutierrez N, Park S-W, Wu J, Lee Y, Suzuki K, et al. Correction of a pathogenic gene mutation in human embryos. Nature. 2017;548(7668):413–9.
How to respond to CRISPR babies. Nature. 2018;564(7734):5–5.
Opar A. CRISPR-edited babies arrived, and regulators are still racing to catch up. Nat Med. 2019;1–3.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Dr. Flannigan reports grants from American Society of Reproductive Medicine, grants from Canadian Urological Association Scholarship Foundation, grants from Canadian Institute for Health Research, grants from Vancouver Coastal Health Research Institute, and grants from New Frontiers Research Fund, outside the submitted work. Dr. Cina and Dr. Phillips have nothing to disclose.
Human and Animal Rights
This article does not contain any studies with human or animal subjects performed by any of the authors.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
This article is part of the Topical Collection on Cell Behavior Manipulation
Rights and permissions
About this article
Cite this article
Cinà, D.P., Phillips, D. & Flannigan, R. CRISPR/Cas9 in Male Factor Infertility. Curr. Tissue Microenviron. Rep. 1, 89–97 (2020). https://doi.org/10.1007/s43152-020-00011-y
Published:
Issue Date:
DOI: https://doi.org/10.1007/s43152-020-00011-y