Abstract
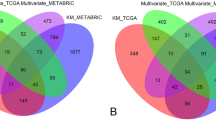
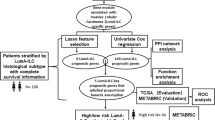
Breast cancer (BRCA) is the most widespread malignant tumor and the leading cause of death in women. BRCA treatments vary based on the presence of estrogen receptors (ER). Generally, cancer preventive and therapy options for ER-negative BRCA are limited compared to ER-positive BRCA. Therefore, this study investigates the key genes predicting poor prognosis for ER-negative BRCA through the application of LASSO (Least Absolute Shrinkage and Selection Operator) and bioinformatics analysis. These methods are analyzed using a dataset GSE7390 that contains 198 untreated lymph node-negative BRCA patients with gene expressions and clinical information. Differentially Expressed Genes (DEGs) between ER-negative and ER-positive BRCA are found using GEO2R. Here, the regularized regression reduces the dimensionality of DEGs by selecting necessary genes. Later, Support Vector Machines for Survival and Random Survival Forest methods were implemented to construct survival predictive models. Comparatively, LASSO holds higher C-index and AUC values with better prediction accuracy and it selects 28 DEGs consisting of 14 upregulated and 14 downregulated genes. Among them, LCN2 expression is significantly downregulated and ZMYND10 expression is significantly upregulated in tumor tissues compared to normal tissue using GEPIA2. Also, both genes are statistically significant in survival rates for TCGA BRCA patients (p < 0.05). Hence, this study highlights higher LCN2 and low ZMYND10 expression in ER-negative BRCA associated with a poor prognosis. Consequently, LCN2 and ZMYND10 genes can be used to predict and serve as potential biomarkers for future diagnosis and treatment in ER-negative BRCA.








Similar content being viewed by others
References
Altman DG, De Stavola BL, Love SB, Stepniewska KA (1995) Review of survival analyses published in cancer journals. Br J Cancer 72(2):511–518. https://doi.org/10.1038/bjc.1995.364
Asri H, Mousannif H, Al Moatassime H, Noel T (2016) Using machine learning algorithms for breast cancer risk prediction and diagnosis. Procedia Comput Sci 83:1064–1069. https://doi.org/10.1016/j.procs.2016.04.224
Barrett T, Wilhite SE, Ledoux P, Evangelista C, Kim IF, Tomashevsky M, Marshall KA, Phillippy KH, Sherman PM, Holko M, Yefanov A, Lee H, Zhang N, Robertson CL, Serova N, Davis S, Soboleva A (2013) NCBI GEO: archive for functional genomics data sets-update. Nucleic Acids Res 41(D1):D991–D995. https://doi.org/10.1093/nar/gks1193
Benjamini Y, Hochberg Y (1995) Controlling the false discovery rate: a practical and powerful approach to multiple testing. J Roy Stat Soc: Ser B (Methodol) 57(1):289–300
Bühlmann P, Van De Geer S (2011) Statistics for high-dimensional data: methods, theory and applications. Springer Science and Business Media
Chowdhury MZI, Turin TC (2020) Variable selection strategies and its importance in clinical prediction modelling. Fam Med Community Health 8(1). https://doi.org/10.1136/fmch-2019-000262
Cox DR (1972) Regression models and life-tables. J Roy Stat Soc: Ser B (Methodol) 34(2):187–202
Cruz JA, Wishart DS (2006) Applications of machine learning in cancer prediction and prognosis. In: Cancer informatics, vol 2. https://doi.org/10.1177/117693510600200030
Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA (2003) DAVID: database for annotation, visualization, and integrated discovery. Genome Biol 4(9):1–11. https://doi.org/10.1186/gb-2003-4-9-r60
Desmedt C, Piette F, Loi S, Wang Y, Lallemand F, Haibe-Kains B, Viale G, Delorenzi M, Zhang Y, D’Assignies MS, Bergh J, Lidereau R, Ellis P, Harris AL, Klijn JGM, Foekens JA, Cardoso F, Piccart MJ, Buyse M, Sotiriou C (2007) Strong time dependence of the 76-gene prognostic signature for node-negative breast cancer patients in the TRANSBIG multicenter independent validation series. Clin Cancer Res 13(11):3207–3214. https://doi.org/10.1158/1078-0432.CCR-06-2765
Harbeck N, Penault-Llorca F, Cortes J, Gnant M, Houssami N, Poortmans P, Ruddy K, Tsang J, Cardoso F (2019) Breast cancer. Nat Rev Dis Primers 5:66. https://doi.org/10.1038/s41572-019-0111-2
Hoerl AE, Kennard RW (1970) Ridge regression: Biased estimation for nonorthogonal problems. Technometrics 12(1):55–67. https://doi.org/10.1080/00401706.1970.10488634
Huang DW, Sherman BT, Lempicki RA (2009) Systematic and integrative analysis of large gene lists using DAVID bioinformatics resources. Nat Protoc 4(1):44–57. https://doi.org/10.1038/nprot.2008.211
Ikeda K, Horie-Inoue K, Inoue S (2015) Identification of estrogen-responsive genes based on the DNA binding properties of estrogen receptors using high-throughput sequencing technology. Acta Pharmacol Sin 36(1):24–31. https://doi.org/10.1038/aps.2014.123
Ishwaran H, Kogalur UB (2019) Fast unified random forests for survival, regression, and classification (RF-SRC). R package version 2(1)
Ishwaran H, Kogalur UB, Blackstone EH, Lauer MS (2008) Random survival forests. Ann Appl Stat 2(3). https://doi.org/10.1214/08-AOAS169
Johnstone IM, Titterington DM (2009) Statistical challenges of high-dimensional data. Philos Trans R Soc A: Math Phys Eng Sci 367(1906):4237–4253. https://doi.org/10.1098/rsta.2009.0159
Kanehisa M, Goto S (2000) KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28(1):27–30. https://doi.org/10.1093/nar/28.1.27
Kim H, Park T, Jang J, Lee S (2022) Comparison of survival prediction models for pancreatic cancer: Cox model versus machine learning models. Genomics Inform 20(2). https://doi.org/10.5808/gi.22036
Kourou K, Exarchos TP, Exarchos KP, Karamouzis MV, Fotiadis DI (2015) Machine learning applications in cancer prognosis and prediction. Comput Struct Biotechnol J 13:8–17. https://doi.org/10.1016/j.csbj.2014.11.005
Lin RH, Lin CS, Chuang CL, Kujabi BK, Chen YC (2022) Breast cancer survival analysis model. Appl Sci 12(4):1971. https://doi.org/10.3390/app12041971
Litzenburger BC, Brown PH (2014) Advances in preventive therapy for estrogen-receptor-negative breast cancer. Curr Breast Cancer Rep 6:96–109. https://doi.org/10.1007/s12609-014-0144-1
Newson R (2006) Confidence intervals for rank statistics: Somers’ D and extensions. Stand Genomic Sci 6(3):309–334. https://doi.org/10.1177/1536867x0600600302
Omurlu IK, Ture M, Tokatli F (2009) The comparisons of random survival forests and Cox regression analysis with simulation and an application related to breast cancer. Expert Syst Appl 36(4):8582–8588. https://doi.org/10.1016/j.eswa.2008.10.023
Pölsterl S, Navab N, Katouzian A (2015) Fast training of support vector machines for survival analysis. In: Machine learning and knowledge discovery in databases: European conference, ECML PKDD 2015, Porto, Portugal, September 7–11, 2015, Proceedings, Part II 15, pp 243–259. Springer International Publishing. https://doi.org/10.1007/978-3-319-23525-7_15
Putti TC, Abd El-Rehim DM, Rakha EA, Paish CE, Lee AH, Pinder SE, Ellis IO (2005) Estrogen receptor-negative breast carcinomas: a review of morphology and immunophenotypical analysis. Mod Pathol 18(1):26–35. https://doi.org/10.1038/modpathol.3800255
Reis-Filho JS, Pusztai L (2011) Gene expression profiling in breast cancer: classification, prognostication, and prediction. The Lancet 378(9805):1812–1823. https://doi.org/10.1016/S0140-6736(11)61539-0
Ritchie ME, Phipson B, Wu DI, Hu Y, Law CW, Shi W, Smyth GK (2015) limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res 43(7):e47. https://doi.org/10.1093/nar/gkv007
Shivaswamy PK, Chu W, Jansche M (2007) A support vector approach to censored targets. In: Seventh IEEE international conference on data mining (ICDM), pp 655–660. https://doi.org/10.1109/ICDM.2007.93
Statnikov A, Wang L, Aliferis CF (2008) A comprehensive comparison of random forests and support vector machines for microarray-based cancer classification. BMC Bioinformatics 9(1):1–10. https://doi.org/10.1186/1471-2105-9-319
Szklarczyk D, Gable AL, Lyon D, Junge A, Wyder S, Huerta-Cepas J, Simonovic M, Doncheva NT, Morris JH, Bork P, Jensen LJ, Mering CV (2019) STRING v11: protein–protein association networks with increased coverage, supporting functional discovery in genome-wide experimental datasets. Nucleic Acids Res 47(D1):D607–D613. https://doi.org/10.1093/nar/gky1131
Tang Z, Li C, Kang B, Gao G, Li C, Zhang Z (2017) GEPIA: a web server for cancer and normal gene expression profiling and interactive analyses. Nucleic Acids Res 45(W1):W98–W102. https://doi.org/10.1093/nar/gkx247
Tang Z, Kang B, Li C, Chen T, Zhang Z (2019) GEPIA2: an enhanced web server for large-scale expression profiling and interactive analysis. Nucleic Acids Res 47(W1):W556–W560. https://doi.org/10.1093/nar/gkz430
Tibshirani R (1996) Regression shrinkage and selection via the lasso. J R Stat Soc Ser B Stat Methodol 58(1):267–288. https://doi.org/10.1111/j.2517-6161.1996.tb02080.x
Tibshirani R (1997) The lasso method for variable selection in the Cox model. Stat Med 16(4):385–395
Turner NC, Neven P, Loibl S, Andre F (2017) Advances in the treatment of advanced oestrogen-receptor-positive breast cancer. The Lancet 389(10087):2403–2414. https://doi.org/10.1016/S0140-6736(16)32419-9
Uchida S, Sugino T (2022) In silico identification of genes associated with breast cancer progression and prognosis and novel therapeutic targets. Biomedicines 10(11):2995. https://doi.org/10.3390/biomedicines10112995
Uno H, Cai T, Tian L, Wei LJ (2007) Evaluating prediction rules for t-year survivors with censored regression models. J Am Stat Assoc 102(478):527–537. https://doi.org/10.1198/016214507000000149
Usman M, Doguwa SIS, Alhaji BB (2021) Comparing the prediction accuracy of Ridge, Lasso and Elastic Net regression models with linear regression using breast cancer data. Bayero J Pure Appl Sci 14(2):134–149. https://doi.org/10.4314/bajopas.v14i2.16
Van Belle V, Pelckmans K, Van Huffel S, Suykens JA (2011) Support vector methods for survival analysis: a comparison between ranking and regression approaches. Artif Intell Med 53(2):107–118. https://doi.org/10.1016/j.artmed.2011.06.006
Wang H, Li G (2017) A selective review on random survival forests for high dimensional data. Quant Bio-Sci 36(2):85. https://doi.org/10.22283/qbs.2017.36.2.85
**ao J, Mo M, Wang Z, Zhou C, Shen J, Yuan J, He Y, Zheng Y (2022) The application and comparison of machine learning models for the prediction of breast cancer prognosis: retrospective cohort study. JMIR Med Inform 10(2):e33440. https://doi.org/10.2196/33440
Yu SH, Cai JH, Chen DL, Liao SH, Lin YZ, Chung YT, Tsai JJP, Wang CC (2021) LASSO and bioinformatics analysis in the identification of key genes for prognostic genes of gynecologic cancer. J Pers Med 11(11):1177. https://doi.org/10.3390/jpm11111177
Zemmour C, Bertucci F, Finetti P, Chetrit B, Birnbaum D, Filleron T, Boher JM (2015) Prediction of early breast cancer metastasis from DNA microarray data using high-dimensional cox regression models. Cancer Inform 14:CIN-S17284. https://doi.org/10.4137/CIN.S17284
Zhang Y, Wong G, Mann G, Muller S, Yang JY (2022) SurvBenchmark: comprehensive benchmarking study of survival analysis methods using both omics data and clinical data. GigaScience 11. https://doi.org/10.1093/gigascience/giac071
Zou H, Hastie T (2005) Regularization and variable selection via the elastic net. J R Stat Soc Ser B Stat Methodol 67(2):301–320. https://doi.org/10.1111/j.1467-9868.2005.00503.x
Funding
No funds were received for this research.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
The authors declared that there is no conflict of interest regarding the publication of this paper.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Divya, P., Suresh, S. Bioinformatics Analysis in the Identification of Prognostic Signatures for ER-Negative Breast Cancer Data. J Indian Soc Probab Stat 25, 1–16 (2024). https://doi.org/10.1007/s41096-024-00187-8
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s41096-024-00187-8
Keywords
- Breast cancer
- Estrogen receptors
- Survival analysis
- Regularized regression
- Machine learning survival methods
- Bioinformatics analysis