Abstract
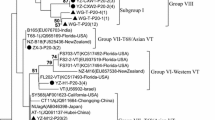
Citrus tristeza virus (CTV) is a viral pathogen that affects the citrus industry worldwide. Knowledge of the genetic structure and evolution of CTV, particularly in CTV from wild mandarins, is limited but essential in develo** sustainable management strategies. With CTV isolates collected in the Nanling Mountains of China, the population structure and genetic evolution among populations were analyzed in this study. The CP/Hinf I RFLP analysis revealed many mild strains, and RFLP group III is the dominant group in the Nanling Mountains of China. Genotypes VT, T3, T30, T36, B165, HA, and RB were detected with VT, and T3 were observed in high frequencies, indicating that the CTV population structure was complex and widely diverse. In the phylogenetic trees of CP, p20, and p23 genes, CTV isolates from Chongyi (M-CY), Yizhang (M-YZ), and Daoxian (M-DX) tended to group, indicating that CTV diversification was maintained by geography-driven. The nucleotide diversity (Pi) value showed the highest genetic diversity observed in populations from Jiangxi (JX) and Hunan (HN), which was consistent with the result of phylogenetic trees. The value of natural selection in all CTV populations was less than 1, implying purifying selection played an essential role in the maintenance of genetic diversity. The neutrality tests and mismatch distribution results suggested that CTV populations were relatively stable. This study provided important information for a better understanding of the evolution of the CTV population.



Similar content being viewed by others
Data availability
We declared that materials described in the manuscript, including all relevant raw data, will be freely available to any scientist wishing to use them for non-commercial purposes, without breaching participant confidentiality.
References
Albiach-Marti MR, Mawassi M, Gowda S, Satyanarayana T, Hilf ME, Shanker S, Almira EC, Vives MC, Lopez C, Guerri J, Flores R, Moreno P, Garnsey SM, Dawson WO (2000) Sequences of citrus tristeza virus separated in time and space are essentially identical. Journal of Virology 74:6856–6865
Albiach-Marti MR, Robertson C, Gowda S, Tatineni S, Belliure B, Garnsey SM, Folimonova SY, Moreno P, Dawson WO (2010) The pathogenicity determinant of citrus tristeza virus causing the seedling yellows syndrome maps at the 3’-terminal region of the viral genome. Molecular Pant Pathology 11:55–67
Ambros S, Gomez-Munoz N, Gimenez-Santamarina S, Sanchez-Vicente J, Navarro-Lopez J, Martinez F, Daros JA, Rodrigo G (2021) Molecular signatures of silencing suppression degeneracy from a complex RNA virus. PLoS Computational Biology 17:e1009166
Baba VY, Giampani JS, Tazima ZH, Yada IFU, Paccola-Meirelles LD, Leite Júnior RP (2014) Agronomic performance of Pera and related sweet orange accessions naturally infected with citrus tristeza virus in northern Paraná State, Brazil. Tropical Plant Pathology 39:442–448
Bar-Joseph M, Marcus R, Lee RF (1989) The continuous challenge of citrus tristeza virus control. Annual Review of Phytopathology 27:291–316
Bederski K, Roistacher CN, Silvestre OP, Müller GW (2010) Long-term cross protection of severe stem pitting citrus tristeza virus in Peru. International Organization of Citrus Virologists Conference Proceedings 1957–2010(17):67–79
Biswas KK, Tarafdar A, Diwedi S, Lee RF (2012) Distribution, genetic diversity and recombination analysis of citrus tristeza virus of India. Virus Genes 45:139–148
Chapman EJ, Prokhnevsky AI, Gopinath K, Dolja VV, Carrington JC (2004) Viral RNA silencing suppressors inhibit the microRNA pathway at an intermediate step. Genes & Development 18:1179–1186
Chen AYS, Peng JHC, Polek M, Tian T, Ludman M, Fátyol K, Ng JCK (2020) Comparative analysis identifies amino acids critical for citrus tristeza virus (T36CA) encoded proteins involved in suppression of RNA silencing and differential systemic infection in two plant species. Molecular Plant Pathology 22:64–76
Chen Y, You X, Chen B, Yi L (2021) Genetic characteristics of citrus tristeza virus isolates from cultivated citrus in China based on coat protein gene. Journal of Biosciences and Medicines 09:190–200
Datta S, Das B, Gopalakrishnan R, Muaka V, Meghvansi MK, Vairale MG, Rahman S, Dwivedi SK, Veer V (2021) Detection of ‘ancestral’ western lineage of citrus tristeza virus virulent genotype in declining Arunachal Wakro orange. Tropical Plant Pathology 46:493–505
Davino S, Panno S, Rangel EA, Davino M, Bellardi MG, Rubio L (2012) Population genetics of cucumber mosaic virus infecting medicinal, aromatic and ornamental plants from northern Italy. Archives of Virology 157:739–745
Davino S, Willemsen A, Panno S, Davino M, Catara A, Elena SF, Rubio L (2013) Emergence and phylodynamics of citrus tristeza virus in Sicily. Italy Plos One 8:e66700
Elhaddad A, ElAmrani A, Fereres A, Moreno A (2016) Spatial and temporal spread of citrus tristeza virus and its aphid vectors in the North western area of Morocco. Insect Science 23:903–912
Excoffier L, Laval G, Schneider S (2017) Arlequin (version 3.0): an integrated software package for population genetics data analysis. Evolutionary Bioinformatics 1:1–47
Folimonova SY (2020) Citrus tristeza virus: a large RNA virus with complex biology turned into a valuable tool for crop protection. PLoS Pathogens 16:e1008416
Fontana A, Debreczeni DE, Albanese G, Davino S, Flores R, Rubio L (2014) Evolutionary analysis of citrus tristeza virus outbreaks in Calabria, Italy: two rapidly spreading and independent introductions of mild and severe isolates. European Journal of Plant Pathology 140:607–613
Gao F, Du Z, Shen J, Yang H, Liao F (2018) Genetic diversity and molecular evolution of ornithogalum mosaic virus based on the coat protein gene sequence. PeerJ 6:e4550
Gillings M, Broadbent P, Indsto J, Lee R (1993) Characterisation of isolates and strains of citrus tristeza closterovirus using restriction analysis of the coat protein gene amplified by the polymerase chain reaction. Journal of Virological Methods 44:305–317
Harper SJ (2013) Citrus tristeza virus: evolution of complex and varied genotypic groups. Frontiers in Microbiology 4:93
Hsieh WH, Chen YC, Liao HC, Lin YR, Chen CH (2021) High differentiation among populations of green foxtail, setaria viridis, in Taiwan and adjacent islands revealed by microsatellite markers. Diversity 13:159
Jiang B, Hong N, Wang GP, Hu J, Zhang JK, Wang CX, Liu Y, Fan XD (2008) Characterization of citrus tristeza virus strains from southern China based on analysis of restriction patterns and sequences of their coat protein genes. Virus Genes 37:185–192
Kavous A, Kamaruzaman S, Ganesan V, Hawa J (2011) Molecular characterization of citrus tristeza virus strains in Peninsular Malaysia. African Journal of Microbiology Research 5:2838–2846
Kumar S, Stecher G, Li M, Knyaz C, Tamura K (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution 35:1547–1549
Librado P, Rozas J (2009) DnaSP v5: a software for comprehensive analysis of DNA polymorphism data. Bioinformatics 25:1451–1452
Liu Q, Zhang S, Mei S, Zhou Y, Wang J, Han GZ, Chen L, Zhou C, Cao M (2021) Viromics unveils extraordinary genetic diversity of the family Closteroviridae in wild citrus. PLoS Pathogens 17:e1009751
López C, Ayllón M, Navas-Castillo J, Guerri J, Moreno P, Flores R (1998) Molecular variability of the 5′-and 3′-terminal regions of citrus tristeza virus RNA. Phytopathology 88:685–691
Lu R, Folimonov A, Shintaku M, Li WX, Falk BW, Dawson WO, Ding SW (2004) Three distinct suppressors of RNA silencing encoded by a 20-kb viral RNA genome. Proceedings of the National Academy of Sciences 101:15742–15747
Martin S, Sambade A, Rubio L, Vives MC, Moya P, Guerri J, Elena SF, Moreno P (2009) Contribution of recombination and selection to molecular evolution of citrus tristeza virus. Journal of General Virology 90:1527–1538
Martin DP, Murrell B, Golden M, Khoosal A, Muhire B (2015) RDP4: Detection and analysis of recombination patterns in virus genomes. Virus Evolution 2015(1):eve003
Matos LA, Hilf ME, Cayetano XA, Feliz AO, Harper SJ, Folimonova SY (2013) Dramatic Change in citrus tristeza virus Populations in the Dominican Republic. Plant Disease 97:339–345
Mawassi M, Mietkiewska E, Gofman R, Yang G, Bar-Joseph MJJOGV (1996) Unusual sequence relationships between two isolates of citrus tristeza virus. Journal of Virology 77:2359–2364
Moreno AB, López-Moya JJ (2020) When viruses play team sports: mixed infections in plants. Phytopathology 110:29–48
Moreno P, López C, Ruiz-Ruiz S, Pena L, Guerri J (2022) From the smallest to the largest subcellular plant pathogen: citrus tristeza virus and its unique p23 protein. Virus Research 314:198755
Pantaleo V, Szittya G, Burgyan J (2007) Molecular bases of viral RNA targeting by viral small interfering RNA-programmed RISC. Journal of Virology 81:3797–3806
Park JW, da Graca JV, Setamou M, Kunta M (2021) Diversity of citrus tristeza virus Strains in the Upper Gulf Coast Area of Texas. Plant Disease 105:592–598
Read DA, Pietersen G (2015) Genotypic diversity of citrus tristeza virus within red grapefruit, in a field trial site in South Africa. European Journal of Plant Pathology 142:531–545
Roy A, Ananthakrishnan G, Hartung JS, Brlansky RH (2010) Development and application of a multiplex reverse-transcription polymerase chain reaction assay for screening a global collection of citrus tristeza virus isolates. Phytopathology 100:1077–1088
Rubio L, Ayllon MA, Kong P, Fernandez A, Polek M, Guerri J, Moreno P, Falk BW (2001) Genetic variation of citrus tristeza virus isolates from California and Spain: evidence for mixed infections and recombination. Journal of Virology 75:8054–8062
Rubio L, Galipienso L, Ferriol I (2020) Detection of plant viruses and disease management: relevance of genetic diversity and evolution. Frontiers in Plant Science 11:1092
Sagheer A, Yongqing L, Mengji C, Fang yun Y, Yan Z, Changyong Z (2011) Molecular characterization of citrus tristeza virus isolates from Pakistan based on CPG/Hinf I restriction fragment length polymorphism (RFLP) groups analysis. African Journal of Biotechnology 10:8689–8694
Sambade A, Rubio L, Garnsey SM, Costa N, Müller GW, Peyrou M, Guerri J, Moreno P (2002) Comparison of viral RNA populations of pathogenically distinct isolates of citrus tristeza virus: application to monitoring cross-protection. Plant Pathology 51:257–265
Scott KA, Hlela Q, Zablocki O, Read D, van Vuuren S, Pietersen G (2012) Genotype composition of populations of grapefruit-cross-protecting citrus tristeza virus strain GFMS12 in different host plants and aphid-transmitted sub-isolates. Archives of Virology 158:27–37
Semorile L, Gago-Zachert S, Costa N, Grau O (1999) Sequence variability in p27 gene of citrus tristeza virus (CTV) revealed by SSCP analysis. Electronic Journal Of Biotechnology 2:3–4
Sentandreu V, Castro JA, Ayllón MA, Rubio L, Guerri J, González-Candelas F, Moreno P, Moya A (2006) Evolutionary analysis of genetic variation observed in citrus tristeza virus (CTV) after host passage. Archives of Virology 151:875–894
Shilts T, El-Mohtar C, Dawson WO, Killiny N (2020) citrus tristeza virus P33 protein is required for efficient transmission by the Aphid Aphis (Toxoptera) citricidus (Kirkaldy). Viruses 12:1131
Silva G, Marques N, Nolasco G (2012) The evolutionary rate of citrus tristeza virus ranks among the rates of the slowest RNA viruses. Journal of General Virology 93:419–429
Singh JK, Tarafdar A, Sharma SK, Biswas KK (2013) Evidence of recombinant citrus tristeza virus isolate occurring in acid lime cv. pant lemon orchard in Uttarakhand Terai region of Northern Himalaya in India. Indian Journal of Virology 24:35–41
Soler N, Plomer M, Fagoaga C, Moreno P, Navarro L, Flores R, Peña L (2012) Transformation of Mexican lime with an intron-hairpin construct expressing untranslatable versions of the genes coding for the three silencing suppressors of citrus tristeza virus confers complete resistance to the virus. Plant Biotechnology Journal 10:597–608
Wang J, Zhou T, Shen P, Zhang S, Cao M, Zhou Y, Li Z (2020) Complete genome sequences of two novel genotypes of citrus tristeza virus infecting Poncirus trifoliata in China. Journal of Plant Pathology 102:903–907
Was G, Bowen BW (1998) Shallow population histories in deep evolutionary lineages of marine fishes: insights from sardines and anchovies and lessons for conservation. Journal of Heredity 89:415–426
Wright S (1990) Evolution in mendelian populations. Genetics 52:241–295
Wu GW, Tang M, Wang GP, ** FY, Yang ZK, Cheng LJ, Hong N (2015) Genetic diversity and evolution of two capsid protein genes of citrus tristeza virus isolates from China. Archives of Virology 160:787–794
**ao C, Yao RX, Li F, Dai SM, Licciardello G, Catara A, Gentile A, Deng ZN (2017) Population structure and diversity of citrus tristeza virus (CTV) isolates in Hunan Province, China. Archives of Virology 162:409–423
Yi L, Zhou CY (2014) Phylogenetic analysis of citrus tristeza virus isolates of wild type citrus in China. Journal of Integrative Agriculture 13:2669–2677
Yi L, Zhou C, Zhou Y, Wang Z, Tang K (2007) Molecular characterization of citrus tristeza virus isolates in Chinese wild type citrus. Scientia Agricultura Sinica 40:932–939
Yokomi R, Selvaraj V, Maheshwari Y, Chiumenti M, Saponari M, Giampetruzzi A, Weng Z, **ong Z, Hajeri S (2018) Molecular and biological characterization of a novel mild strain of citrus tristeza virus in California. Archives of Virology 163:1795–1804
Young SS, Lin SC, Liu MY (2013) Genetic diversity and population structure of two freshwater copepods (Copepoda: Diaptomidae), Neodiaptomus schmackeri (Poppe and Richard, 1892) and Mongolodiaptomus birulai (Rylov, 1922) from Taiwan. Diversity 5:796–810
Zhou C, Hailstones D, Broadbent P, Connor R, Bowyer J (2002) Studies on mild strain cross protectio against stem-pitting tristeza virus. International Organization of Citrus Virologists Conference Proceedings 1957–2010(15):151–157
Zhou XK, Zhao JF, Wang Y, Czo MJ, Chen CX, Zhou CY, Zhou Y (2022) Molecular characterization of four citrus tristeza virus isolates that cause severe symptoms in navel orange in Ganzhou, China. Tropical Plant Pathology 47:509–520
Funding
This research was supported by project 31860488 from the National Natural Science Foundation of China.
Author information
Authors and Affiliations
Contributions
Field work and specimen collections were carried out by all authors. C.Y., W.C., and Z.K. carried out the experiments. Molecular analyses were carried out by C.Y., W.C., Z.K., and C.B. Manuscript drafting and editing was performed by C.Y. and Y.L. and review and editing by S.L. All authors have read and agreed to the published version of the manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Chen, Y., Yi, L., Zhong, K. et al. Population genetic characteristics of citrus tristeza virus from wild mandarins in the Nanling Mountains of China. Trop. plant pathol. 48, 270–282 (2023). https://doi.org/10.1007/s40858-023-00567-8
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40858-023-00567-8