Abstract
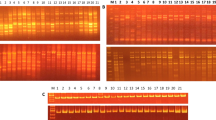
Characterizing the various genotypes of watermelons (Citrullus lanatus) is an important step to manage effectively the use of genotype resources. The aim of this work is to determine the level of genetic diversity among different landraces of Palestinian watermelon using RAPD molecular markers. For this, 14 RAPD primers were used to amplify the DNA extracted from 8 watermelon landraces. The banding pattern from RAPD analysis was scored visually for each primer and the data matrix was developed for UPGMA-based cluster analysis. Using the Nei and Li distance coefficients, cluster analysis was performed. Marker index and polymorphic information content as well as resolving power was calculated for all the markers. Twelve RAPD markers produced various banding patterns and revealed polymorphic bands of which 52 (94.6%) bands were polymorphic and 3 (0.035%) were monomorphic. The highest percentage of polymorphic markers was 100% and the lowest was 60%. The genotypes were mainly divided into two clusters based on the dendrogram derived from RAPD data. Cluster-I, which has two sub-clusters, included seven genotypes, sub-cluster-I.a contained only two genotypes (UB-199–11 and UB-510–20), and sub-cluster-I.b included two groups containing in which genotypes UB-402–19 and UB-578–21-2 represented in group 1 and genotypes UB-307–18-2, UB-377–19, and UB-578–21-1 represented in group 2. Cluster-II contained only one genotype accession (UB-307–18-1). Based on 12 RAPD, the similarity coefficients of Watermelon genotypes ranged from 0.31 to 0.82.



Similar content being viewed by others
Availability of Data and Materials
All data and materials of the study have been presented in the main manuscript.
Abbreviations
- FAO:
-
Food and Agriculture Organization
- AFLP:
-
Amplified fragment length polymorphism
- SSR:
-
Simple sequence repeat
- EST-SSR:
-
Expressed sequence tags-SSRs
- SNP:
-
Single nucleotide polymorphism
- RAPD:
-
Random amplified polymorphic DNA
- UWAC:
-
Union of Agricultural Work Committees
- PCR:
-
Polymerase chain reaction
- UPGMA:
-
Unweighted Pair Group Method with Arithmetic Average
- MVSP:
-
Multi variate statistical package
- MI:
-
Marker index
- PIC:
-
Polymorphic information content
- RP:
-
Resolving power
- NB:
-
Number of bands
- NPB:
-
Number of polymorphic bands
- NMB:
-
Number of monomorphic bands
- PPB:
-
Percentage of polymorphic bands
References
Meeuse ADJ (1962) The cucurbitaceae of southern Africa. Bothalia 8(1):1–111
Whitaker TW, Bemis WP (1975) Origin and evolution of the cultivated cucurbita. Bull Torrey Bot Club 102(6):362–368. https://doi.org/10.2307/2484762
Zamir D, Navot N, Rudich J (1984) Enzyme polymorphisms in Citrullus lanatus and Citrullus colocynthis in Israel and Sinai. Plant Syst Evol 146(163):170
Jarret RL, Merrick LC, Holms T, Evans J, Aradhya MK (1997) Simple sequence repeats in watermelon (Citrullus lanatus (Thunb.) Matsum. & Nakai). Genome 40(433):441
Tamburini E, Costa S, Rugiero I, Pedrini P, Marchetti MG (2017) Quantification of Lycopene, β-carotene, and total soluble solids in intact red-flesh watermelon (Citrullus lanatus) using on-line near-infrared spectroscopy. Sensors 17(4):746. https://doi.org/10.3390/s17040746
Makaepea M, Beswa MD, Jideani AIO (2019) Watermelon as a potential fruit snack. Int J Food Prop 22(1):355–370. https://doi.org/10.1080/10942912.2019.1584212
Edwards AJ, Vinyard BT, Wiley ER, Brown ED, Collins JK, Perkins-Veazie P, Baker RA, Clevidence BA (2003) Consumption of watermelon juice increases plasma concentrations of lycopene and beta-carotene in humans. J Nutr 133(4):1043–1050. https://doi.org/10.1093/jn/133.4.1043. (PMID: 12672916)
Naz A, Butt MS, Sultan MT, Qayyum MMN, Niaz RS (2014) Watermelon lycopene and allied health claims. Excli J 13:650–666
Lum T, Connolly M, Marx A, Beidler J, Hooshmand S, Kern M, Liu C, Hong MY (2019) Effects of fresh watermelon consumption on the acute satiety response and cardiometabolic risk factors in overweight and obese adults. Nutrients 11(3):595. https://doi.org/10.3390/nu11030595
Food and agriculture organization of the United Nations (2020) FAOSTAT statistical database. [Rome]: FAO
PCBS (2021). Database of the Palestinian Central Bureau of statistics. Available at https://www.pcbs.gov.ps/pcbs_2012/Publications_AR.aspx
Alimari A, Zaid A, Fadda Z (2017) Genetic diversity in local Palestinian watermelon (Citrullus lanatus) accessions. Int J Agric Policy Res 5(10):157–162
Yang X, Ren R, Ray R, Xu J, Li P, Zhang M, Kilian A (2016) Genetic diversity and population structure of core watermelon (Citrullus lanatus) genotypes using DArTseq-based SNPs. Plant Genetic Resour 14(3):226–233. https://doi.org/10.1017/S1479262115000659
Powell W, Morgante M, Andre C, Hanafey M, Vogel J, Tingey S, Rafalski A (1996) The comparison of RFLP, RAPD, AFLP and SSR (microsatellite) markers for genotype analysis. Mol Breeding 2:225–238
Che KP, Liang CY, Wang YG, ** DM, Wang B, Xu Y, Kang GB, Zhang HY (2003) Genetic assessment of watermelon genotype using the AFLP technique. HortScience 38:81–84
Kwon YS, Oh YH, Yi SI, Kim HY, An JM, Yang SG, Ok SH, Shin JS (2010) Informative SSR markers for commercial variety discrimination in watermelon (Citrullus lanatus). Genes Genomics 32:115–122
Mujaju C, Werlemark G, Garkava-Gustavsson L, Nybom H (2010) High Levels of RAPD and SSR marker diversity in landraces of watermelon (Citrullus lanatus) in Southern Africa. Acta Hort 918:291–296
Mujaju C, Sehic J, Nybom H (2013) Assessment of EST-SSR markers for evaluating genetic diversity in watermelon accessions from Zimbabwe. Am J Plant Sci 4:1448
Sneath PHA, Sokal RR (1973) Numerical taxonomy—the principles and practice of numerical classification. (W. H. Freeman: San Francisco)
Nei M (1978) Estimation of average heterozygosity and genetic distance from a small number of individuals. Genetics 89:583–590
Kovach WL (2007) A multivariate statistical package (MVSP) for windows ver 3.22. Kovach Computing Services, Pentraeth
Prevost A, Wilkinson MJ (1999) A new system of comparing PCR primers applied to ISSR fingerprinting of potato cultivars. Theor Appl Genet 98(1):107–112
Garcia-Mas J, Olivier M, Gomez-Paniagua H, De Vicente MC (2000) Comparing AFLP, RAPD and REFLP markers for measuring genetic diversity in melon. Theor Appl Genet 101:860–864
Kayesh E (2018) Morphological and molecular characterization of watermelon genotypes using RAPD markers. Fund Appl Agri 3(3):573–578
Elbekkay M, Hamza H, Neily MH, Djebali N, Ferchichi A (2021) Characterization of watermelon local cultivars from Southern Tunisia using morphological traits and molecular markers. Euphytica 217:74. https://doi.org/10.1007/s10681-021-02809-9
Lee SJ, Shin JS, Park KW, Hong YP (1996) Detection of genetic diversity using RAPD-PCR and sugar analysis in watermelon [Citrullus lanatus (Thumb.) Mansf.] germplasm. Theor Appl Genet 92:719–725
Levi A, Thomas CE, Wehner TC, Zhang X (2001) Low genetic diversity indicates the need to broaden the genetic base of cultivated watermelon. HortScience 36:1096–1101
Levi A, Thomas CE, Keinath AP, Wehner TC (2001) Genetic diversity among watermelon (Citrullus lanatus and Citrullus colocynthis) accessions. Genet Resour Crop Evol 48:559–566
Mujaju C, Sehic J, Werlemark G, Garkava-Gustavsson L, Fatih M, Nybom H (2010) Genetic diversity in watermelon (Citrullus lanatus) landraces from Zimbabwe revealed by RAPD and SSR markers. Hereditas 147:142–153
Solmaz I, Sari N, Aka-Kacar Y, Yalcin-Mendi YN (2010) The genetic characterization of Turkish watermelon (Citrullus lanatus) accessions using RAPD markers. Genet Resour Crop Evolut 57:763–771
Acknowledgements
The authors are thankful to the Union of Agricultural Work Committees (UWAC) for providing the watermelon landraces seeds.
Funding
No funding was received for conducting this study.
Author information
Authors and Affiliations
Contributions
Yamen Hamdan, Abdalmenem Hawamda and Mazen Salman designed and carried out the study. Laith badran, Estabraq Daraghmeh, Linda Mar’i, Anas Khalaf, Ramiz Maraabeh and Raghda Ibrahim carried out the field and laboratory work. Yamen Hamdan and Rezq Basheer-Salimia carried out the statistical analyses and interpretation of the study. Yamen Hamdan and Abdalmenem Hawamda wrote the first draft of the manuscript. Yamen Hamdan wrote the final draft of manuscript. Yamen Hamdan and Mazen Salman reviewed the final draft. The authors read and approved the final manuscript.
Corresponding author
Ethics declarations
Competing interests
The authors declare that they have no competing interests.
Consent for Publication
The authors declare that the work has been consented for publication.
Ethics Approval
Not applicable.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance Statement: Characterizing watermelon genotype is an important step for effective management of genetic resources. Utilizing molecular methods, this study provide valuable insights into the genetic diversity of Palestinian watermelon landraces, essential for conservation and breeding ctivities.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Hamdan, Y.A.S., Hawamda, A.I.M., badran, L. et al. Exploring Genetic Variations Among Palestinian Watermelon (Citrullus lanatus) Germplasm Using RAPD Molecular Markers. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. (2024). https://doi.org/10.1007/s40011-024-01612-5
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s40011-024-01612-5