Abstract
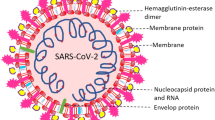
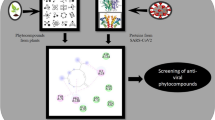
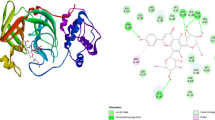
The fatal virus SARS-CoV-2 emerged in December 2019, which caused a global pandemic of coronavirus disease, also known as COVID-19. Identification of the naturally occurring bioactive compounds that can inhibit or modulate the proteins and enzymes of the virus SARS-CoV-2 by targeting the host cells and immune system of the human body is an instrumental approach for COVID-19 therapy. It is an attempt to assess the potential of naturally occurring bioactive compounds and their derivatives against coronavirus using molecular docking and molecular dynamics simulation studies, i.e. binding affinity of the active receptors of the host cells to the selected bioactive compound against SARS-CoV-2. Docking and MD simulation studies were carried out in this study to estimate the affinity of the selected bioactive compounds along with the reference drugs remdesivir and ivermectin in opposition to the vital target proteins and enzymes of coronavirus. Among all selected bioactives, as per the computational docking parameters, curcumin has shown excellent binding, especially against ACE-2 and RdRp. Apigenin also showed encouraging results against spike protein comparable to reference drugs remdesivir and ivermectin. These results are furthermore established by the molecular dynamic simulation study and analysis of protein–ligand complex, which shows minimum binding deviation and minor average fluctuations in energies of protein residues throughout the simulation period. Curcumin glucuronide and apigenin 7-glucoside showed the most suitable response as they have a marvellous binding affinity with COVID-19 targets that are responsible for viral entry as well as their further replication into the host cell; hence, they could be some possible potential drug candidates for COVID-19 therapy.






Similar content being viewed by others
Data Availability
The authors confirm that the data supporting the findings of the study are available within the article and its supplementary materials.
References
Sachs JD, Horton R, Bagenal J et al (2020) The Lancet COVID-19 Commission. Lancet 396:454–455. https://doi.org/10.1016/S0140-6736(20)31494-X
Zhou Y, Hou Y, Shen J et al (2020) Network-based drug repurposing for novel coronavirus 2019-nCoV/SARS-CoV-2. Cell Discov 6:14. https://doi.org/10.1038/s41421-020-0153-3
Eweas AF, Alhossary AA, Abdel-Moneim AS (2020) Molecular docking reveals ivermectin and remdesivir as potential repurposed drugs against SARS-CoV-2. Front Microbiol 11:592908. https://doi.org/10.3389/fmicb.2020.592908
Bajorath J (2002) Integration of virtual and high-throughput screening. Nat Rev Drug Discov 1:882–894. https://doi.org/10.1038/nrd941
Soni U, Singh P, Gupta OP et al (2022) Lichen planus drugs re-purposing as potential anti COVID-19 therapeutics through molecular docking and molecular dynamics simulation approach. J Clin Transl Res 8:127–146. https://doi.org/10.18053/jctres.08.202202.003
Mollica V, Rizzo A, Massari F (2020) The pivotal role of TMPRSS2 in coronavirus disease 2019 and prostate cancer. Future Oncol 16:2029–2033. https://doi.org/10.2217/fon-2020-0571
Gao Y, Yan L, Huang Y et al (2020) Structure of the RNA-dependent RNA polymerase from COVID-19 virus. Science 368:779–782. https://doi.org/10.1126/science.abb7498
Kim K-J, Liu X, Komabayashi T et al (2016) Natural products for infectious diseases. Evid Based Complement Altern Med 2016:9459047. https://doi.org/10.1155/2016/9459047
Chakraborty C, Sharma AR, Mallick B et al (2021) Evaluation of molecular interaction, physicochemical parameters and conserved pattern of SARS-CoV-2 Spike RBD and hACE2: in silico and molecular dynamics approach. Eur Rev Med Pharmacol Sci 25:1708–1723. https://doi.org/10.26355/eurrev_202102_24881
Almeida-Neto FWQ, Castro Matos MG, Marinho EM et al (2021) In silico study of the potential interactions of 4’-acetamidechalcones with protein targets in SARS-CoV-2. Biochem Biophys Res Commun 537:71–77. https://doi.org/10.1016/j.bbrc.2020.12.074
Parihar A, Sonia ZF, Akter F et al (2022) Phytochemicals-based targeting RdRp and main protease of SARS-CoV-2 using docking and steered molecular dynamic simulation: a promising therapeutic approach for Tackling COVID-19. Comput Biol Med 145:105468. https://doi.org/10.1016/j.compbiomed.2022.105468
Wang S, Fang X, Wang Y (2022) In silico screening of novel TMPRSS2 inhibitors for treatment of COVID-19. Molecules 27:4210. https://doi.org/10.3390/molecules27134210
Domínguez-Villa FX, Durán-Iturbide NA, Ávila-Zárraga JG (2021) Synthesis, molecular docking, and in silico ADME/Tox profiling studies of new 1-aryl-5-(3-azidopropyl)indol-4-ones: potential inhibitors of SARS CoV-2 main protease. Bioorg Chem 106:104497. https://doi.org/10.1016/j.bioorg.2020.104497
Hostetler GL, Ralston RA, Schwartz SJ (2017) Flavones: food sources, bioavailability, metabolism, and bioactivity. Adv Nutr 8:423–435. https://doi.org/10.3945/an.116.012948
Pandareesh MD, Shrivash MK, Naveen Kumar HN et al (2016) Curcumin monoglucoside shows improved bioavailability and mitigates rotenone induced neurotoxicity in cell and Drosophila models of Parkinson’s disease. Neurochem Res 41:3113–3128. https://doi.org/10.1007/s11064-016-2034-6
Maurya VK, Kumar S, Prasad AK et al (2020) Structure-based drug designing for potential antiviral activity of selected natural products from Ayurveda against SARS-CoV-2 spike glycoprotein and its cellular receptor. Virusdisease 31:179–193. https://doi.org/10.1007/s13337-020-00598-8
Fidelis QC, Faraone I, Russo D et al (2019) Chemical and biological insights of Ouratea hexasperma (A. St.-Hil.) Baill.: a source of bioactive compounds with multifunctional properties. Nat Prod Res 33:1500–1503. https://doi.org/10.1080/14786419.2017.1419227
Salehi B, Venditti A, Sharifi-Rad M et al (2019) The therapeutic potential of apigenin. Int J Mol Sci 20:1305. https://doi.org/10.3390/ijms20061305
Villa-Rodriguez JA, Kerimi A, Abranko L et al (2018) Acute metabolic actions of the major polyphenols in chamomile: an in vitro mechanistic study on their potential to attenuate postprandial hyperglycaemia. Sci Rep 8:5471. https://doi.org/10.1038/s41598-018-23736-1
Lim R, Barker G, Wall CA, Lappas M (2013) Dietary phytophenols curcumin, naringenin and apigenin reduce infection-induced inflammatory and contractile pathways in human placenta, foetal membranes and myometrium. Mol Hum Reprod 19:451–462. https://doi.org/10.1093/molehr/gat015
Sharma RA, Steward WP, Gescher AJ (2007) Pharmacokinetics and pharmacodynamics of curcumin. Adv Exp Med Biol 595:453–470. https://doi.org/10.1007/978-0-387-46401-5_20
Acknowledgements
The authors are thankful to the department of Applied Sciences, the Indian Institute of Information Technology, Allahabad, for providing us with the facilities and encouragement.
Funding
This research did not receive any specific grant from funding agencies in the public, commercial, or not-for-profit sectors.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Significance statement: The fatal SARS-CoV-2 emerged in December 2019, which caused a global pandemic of coronavirus disease, also known as COVID-19. Identifying the naturally occurring bioactive compounds that can inhibit or modulate the proteins and enzymes of the virus SARS-CoV-2 by targeting the human body's host cells and the immune system is an instrumental approach for COVID-19 therapy. It is an effort to assess the potential of naturally occurring bioactive compounds and their derivatives against SARS-CoV-2 using molecular docking and molecular dynamics simulation studies. Curcumin has shown excellent binding among all selected bioactive as per the computational docking parameters, especially against ACE-2 and RdRp. Apigenin also showed encouraging results against spike protein comparable to reference drugs remdesivir and ivermectin. These results are confirmed by the molecular dynamics simulation study and analysis of the protein–ligand complex, which shows minimum binding deviation and minor average fluctuations in the energies of protein residues throughout the simulation period. Curcumin glucuronide and apigenin 7-glucoside showed the most suitable response as they have a marvellous binding affinity with SARS-CoV-2 targets responsible for viral entry and their further replication into the host cell; hence, they could be some possible potential drug candidates for COVID-19 therapy.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Soni, U., Mishra, T., Gupta, O.P. et al. In Silico Evaluation of Promising Naturally Occurring Bioactive Ligands Against Molecular Targets of SARS-Cov-2. Proc. Natl. Acad. Sci., India, Sect. B Biol. Sci. 94, 251–260 (2024). https://doi.org/10.1007/s40011-023-01496-x
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s40011-023-01496-x