Abstract
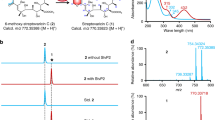
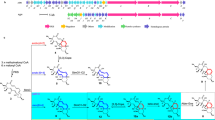
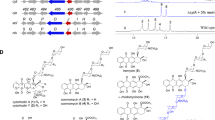
In the present study, an Escherichia coli whole cell system with overexpression of a cytochrome P450 oxidase SlgO1 involved in streptolydigin biosynthetic pathway, an E. coli flavodoxin NADP+ oxidoreductase (EcFLDR), and an E. coli flavodoxin A (EcFLDA) were constructed. Biotransformation experiments revealed that SlgO1 can convert tirandamycin C to tirandamycin F, indicating that it can introduce a hydroxyl group into the C-10 position of tirandamycin C. Subsequently, slgO1 was cloned into pSET152AKE vector under the downstream of ermE* promoter, which was, respectively, introduced into Streptomyces sp. SCSIO1666 (tirandamycin B producer), Streptomyces sp. Ju1008 (tirandamycin C producer), and Streptomyces sp. Ju1009 (tirandamycin E producer). A novel tirandamycin derivative tirandamycin L accumulated in the engineered strain Streptomyces sp. Ju1008::slgO1 was isolated and its structure was determined on the basis of nuclear magnetic resonance (NMR) and mass spectrometry. Unlike most of the identified tirandamycins, tirandamycin L possessed a rare C-11–C-12 saturated bond as well as a C-10 ketone moiety. In addition, tirandamycin L showed weaker antibacterial activity. Based on the structure of tirandamycin L, SlgO1 was proposed to be responsible for multiple modifications toward tirandamycin C, including the formation of C-10 hydroxyl and C-11–C-12 saturated bond.




Similar content being viewed by others
References
Alkhalaf LM, Barry S, Rea D, Gallo A, Griffiths D, Lewandowski J, Fülöp V, Challis GL (2019) Binding of distinct substrate conformations enables hydroxylation of remote sites in thaxtomin D by cytochrome P450 TxtC. J Am Chem Soc 141(1):216–222
Bernhardt R, Urlacher VB (2014) Cytochromes P450 as promising catalysts for biotechnological application: chances and limitations. Appl Microbiol Biotechnol 98(14):6185–6203
Carlson JC, Fortman JL, Anzai Y, Li S, Burr DA, Sherman DH (2010) Identification of the tirandamycin biosynthetic gene cluster from Streptomyces sp. 307-9. ChemBioChem 11(4):564–572
Carlson JC, Li S, Gunatilleke SS, Anzai Y, Burr DA, Podust LM, Sherman DH (2011) Tirandamycin biosynthesis is mediated by co-dependent oxidative enzymes. Nat Chem 3(8):628–633
Cochrane RV, Vederas JC (2014) Highly selective but multifunctional oxygenases in secondary metabolism. Acc Chem Res 47(10):3148–3161
Coon MJ (2005) Cytochrome P450: nature’s most versatile biological catalyst. Annu Rev Pharmacol Toxicol 45:1–25
Duan C, Yao Y, Wang Z, Tian X, Zhang S, Zhang C, Ju J (2010) Fermentation optimization, isolation and identification of tirandamycins A and B from marine-derived Streptomyces sp. SCSIO 1666. Chin J Mar Drugs 29(6):12–20
Girvan HM, Munro AW (2016) Applications of microbial cytochrome P450 enzymes in biotechnology and synthetic biology. Curr Opin Chem Biol 31:136–145
Gómez C, Olano C, Palomino-Schätzlein M, Pineda-Lucena A, Carbajo RJ, Braña AF, Méndez C, Salas JA (2012) Novel compounds produced by Streptomyces lydicus NRRL2433 engineered mutants altered in the biosynthesis of streptolydigin. J Antibiot 65(7):341–348
Lewis DFV, Wiseman A (2005) A selective review of bacterial forms of cytochrome P450 enzymes. Enzym Microb Technol 36(4):377–384
Ma J, Wang Z, Huang H, Luo M, Zuo D, Wang B, Sun A, Cheng YQ, Zhang C, Ju J (2011) Biosynthesis of himastatin: assembly line and characterization of three cytochrome P450 enzymes involved in the post-tailoring oxidative steps. Angew Chem Int Ed 50(34):7797–7802
Macneil DJ, Gewain KM, Ruby CL, Dezeny G, Gibbons PH, Macneil T (1992) Analysis of Streptomyces avermitilis genes required for avermectin biosynthesis utilizing a novel integration vector. Gene 111:61–68
McIver L, Leadbeater C, Campopiano DJ, Baxter RL, Daff SN, Chapman SK, Munro AW (1998) Characterisation of flavodoxin NADP+ oxidoreductase and flavodoxin; key components of electron transfer in Escherichia coli. Eur J Biochem 257(3):577–585
Mo X, Wang Z, Wang B, Ma J, Huang H, Tian X, Zhang S, Zhang C, Ju J (2011a) Cloning and characterization of the biosynthetic gene cluster of the bacterial RNA polymerase inhibitor tirandamycin from marine-derived Streptomyces sp. SCSIO1666. Biochem Biophys Res Commun 406(3):341–347
Mo X, Huang H, Ma J, Wang Z, Wang B, Zhang S, Zhang C, Ju J (2011b) Characterization of TrdL as a 10-hydroxy dehydrogenase and generation of new analogues from an tirandamycin biosynthetic pathway. Org Lett 13(9):2212–2215
Mo X, Ma J, Huang H, Wang Z, Wang B, Zhang S, Zhang C, Ju J (2012) ∆(11,12) double bond formation in tirandamycin biosynthesis is atypically catalyzed by TrdE, a glycoside hydrolase family enzyme. J Am Chem Soc 134(6):2844–2847
Mo X, Li Q, Ju J (2014) Naturally occurring tetramic acid products: isolation, structure elucidation and biological activity. RSC Adv 4:450566–450593
Mo X, Shi C, Gui C, Zhang Y, Ju J, Wang Q (2017) Identification of nocamycin biosynthetic gene cluster from Saccharothrix syringae NRRL B-16468 and generation of new nocamycin derivatives by manipulating gene cluster. Microb Cell Fact 16(1):100
Munro AW, Girvan HM, McLean KJ (2007) Cytochrome P450-redox partner fusion enzymes. Biochim Biophys Acta 1770(3):345–359
Munro AW, Girvan HM, Mason AE, Dunford AJ, McLean KJ (2013) What makes a P450 tick? Trends Biochem Sci 38(3):140–150
Olano C, Gómez C, Pérez M, Palomino M, Pineda-Lucena A, Carbajo RJ, Braña AF, Méndez C, Salas JA (2009) Deciphering biosynthesis of the RNA polymerase inhibitor streptolydigin and generation of glycosylated derivatives. Chem Biol 16(10):1031–1044
Rudolf JD, Chang CY, Ma M, Shen B (2017) Cytochromes P450 for natural product biosynthesis in Streptomyces: sequence, structure, and function. Nat Prod Rep 34(9):1141–1172
**ao J, Zhang Q, Zhu Y, Li S, Zhang G, Zhang H, Saurav K, Zhang C (2013) Characterization of the sugar-O-methyltransferase LobS1 in lobophorin biosynthesis. Appl Microbiol Biotechnol 97(20):9043–9053
Yu Z, Vodanovic-Jankovic S, Ledeboer N, Huang SX, Rajski SR, Kron M, Shen B (2011) Tirandamycins from Streptomyces sp. 17944 inhibiting the parasite Brugia malayi asparagines tRNA synthetase. Org Lett 13(8):2034–2037
Zhang X, Li S (2017) Expansion of chemical space for natural products by uncommon P450 reactions. Nat Prod Rep 34(9):1061–1089
Zhou H, Wang B, Wang F, Yu X, Ma L, Li A, Reetz MT (2019) Chemo- and regioselective dihydroxylation of benzene to hydroquinone enabled by engineered cytochrome P450 monooxygenase. Angew Chem Int Ed Engl 58(3):764–768
Acknowledgements
We are very grateful to Professor Jianhua Ju for providing the tirandamycin-producing strains. Research in this work was supported by Natural Science Foundation of Shandong Province (No. ZR2013CL020) and Research Foundation for Advanced Talents of Qingdao Agricultural University (No. 631301).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
All the authors declare no conflict of interest with respect to this paper.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Mo, X., Gui, C. & Yang, S. Cytochrome P450 oxidase SlgO1 catalyzes the biotransformation of tirandamycin C to a new tirandamycin derivative. 3 Biotech 9, 71 (2019). https://doi.org/10.1007/s13205-019-1611-1
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s13205-019-1611-1