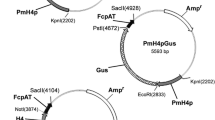
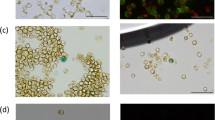
Abstract
The ability to conduct reverse genetic studies in symbiotic systems is enabled by transgene expression and transformation of at least one partner. The symbiotic relationship between the yellow spotted salamander, Ambystoma maculatum, and the green alga, Oophila amblystomatis, is a unique model of vertebrate-algae symbiosis. Despite over 130 years of scientific study, there are still many open questions in this symbiosis. Transgene expression in one partner will accelerate research into the symbiotic relationship. In this paper we describe a tool and method for expression of foreign DNA in, and presumed transformation of, the alga O. amblystomatis. We successfully introduced heritable antibiotic resistance to algal cultures, and observed expression of a green fluorescent reporter protein in all transfected and presumably transformed algal populations. The outcomes of this work enable genetic manipulation of the symbiotic alga O. amblystomatis, allowing direct testing of hypotheses derived from gene expression or genomic studies that will usher in a deeper understanding of the A. maculatum-O. amblystomatis symbiotic system.





Similar content being viewed by others
References
Ahmadjian V (1993) The Lichen symbiosis. John Wiley & Sons, New York
Baier T, Wichmann J, Kruse O, Lauersen KJ (2018) Intron-containing algal transgenes mediate efficient recombinant gene expression in the green microalga Chlamydomonas reinhardtii. Nucleic Acids Res 46:6909–6919. https://doi.org/10.1093/nar/gky532
Bishop C, Jurga E, Graham L (2021) Patterns of bacterial diversity in embryonic capsules of the spotted salamander Ambystoma maculatum: an expanding view of a symbiosis. FEMS Microbiol Ecol 97:fiab128. https://doi.org/10.1093/femsec/fiab128
Burns JA, Zhang H, Hill E et al (2017) Transcriptome analysis illuminates the nature of the intracellular interaction in a vertebrate-algal symbiosis. eLife 6:1–32. https://doi.org/10.7554/elife.22054
Cleves PA, Strader ME, Bay LK et al (2018) CRISPR/Cas9-mediated genome editing in a reef-building coral. Proc Natl Acad Sci 115:5235–5240. https://doi.org/10.1073/pnas.1722151115
Davy SK, Allemand D, Weis VM (2012) Cell biology of cnidarian-dinoflagellate symbiosis. Microbiol Mol Biol Rev 76:229–261. https://doi.org/10.1128/MMBR.05014-11
Decelle J, Colin S, Foster RA (2015) Photosymbiosis in marine planktonic protists. In: Ohtsuka S, Suzaki T, Horiguchi T et al (eds) Marine protists. Springer Japan, Tokyo, pp 465–500
Dunn SR, Phillips WS, Green DR, Weis VM (2007) Knockdown of actin and caspase gene expression by RNA interference in the symbiotic anemone Aiptasia pallida. Biol Bull 212:250–258. https://doi.org/10.2307/25066607
Ellis B, Haal P, Hahne F et al (2022) flowCore: flowCore: Basic structures for flow cytometry data. Version 2.6.0. Bioconductor version: Release (3.14). URL https://bioconductor.org/packages/flowCore/
Endo H, Yoshida M, Uji T et al (2016) Stable nuclear transformation system for the coccolithophorid alga Pleurochrysis carterae. Sci Rep 6:22252. https://doi.org/10.1038/srep22252
Faktorová D, Nisbet RER, Robledo JAF et al (2020) Genetic tool development in marine protists : Emerging model organisms for experimental cell biology. Nat Methods 17(5):481–494. https://doi.org/10.1038/s41592-020-0796-x
Hahne F, Gopalakrishnan N, Khodabakhshi AH et al (2021) flowStats: Statistical methods for the analysis of flow cytometry data. Version 4.6.0. Bioconductor version: Release (3.14). URL http://www.github.com/RGLab/flowStats
Hirakawa Y, Watanabe A (2019) Organellar DNA polymerases in complex plastid-bearing algae. Biomolecules 9:140. https://doi.org/10.3390/biom9040140
Hirakawa Y, Senda M, Fukuda K et al (2021) Characterization of a novel type of carbonic anhydrase that acts without metal cofactors. BMC Biol 19:105. https://doi.org/10.1186/s12915-021-01039-8
Jones VAS, Bucher M, Hambleton EA, Guse A (2018) Microinjection to deliver protein, mRNA, and DNA into zygotes of the cnidarian endosymbiosis model Aiptasia sp. Sci Rep 8:16437. https://doi.org/10.1038/s41598-018-34773-1
Karakashian M (1975) Symbiosis in Paramecium Bursaria. Symposia of the Society for Experimental Biology 145—173. PMID: 785659
Kerney R (2011) Symbioses between salamander embryos and green algae. Symbiosis 54:107–117. https://doi.org/10.1007/s13199-011-0134-2
Kerney R, Leavitt J, Hill E et al (2019) Co-cultures of Oophila amblystomatis between Ambystoma maculatum and Ambystoma gracile hosts show host-symbiont fidelity. Symbiosis 78:73–85. https://doi.org/10.1007/s13199-018-00591-2
Kim B-H, Ramanan R, Cho D-H et al (2012) Simple, rapid and cost-effective method for high quality nucleic acids extraction from different strains of Botryococcus braunii. PLoS ONE 7:e37770. https://doi.org/10.1371/journal.pone.0037770
Kim E, Lin Y, Kerney R et al (2014) Phylogenetic analysis of algal symbionts associated with four North American amphibian egg masses. PLoS ONE 9:e108915. https://doi.org/10.1371/journal.pone.0108915
Kropat J, Hong-Hermesdorf A, Casero D et al (2011) A revised mineral nutrient supplement increases biomass and growth rate in Chlamydomonas reinhardtii. Plant J 66:770–780. https://doi.org/10.1111/j.1365-313X.2011.04537.x
Laemmli UK (1970) Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227:680–685. https://doi.org/10.1038/227680a0
Li X, Patena W, Fauser F et al (2019) A genome-wide algal mutant library and functional screen identifies genes required for eukaryotic photosynthesis. Nat Genet 51:627–635. https://doi.org/10.1038/s41588-019-0370-6
Lo K, Hahne F, Brinkman RR, Gottardo R (2009) flowClust: a Bioconductor package for automated gating of flow cytometry data. BMC Bioinformatics 10:145. https://doi.org/10.1186/1471-2105-10-145
Munakata H, Nakada T, Nakahigashi K et al (2016) Phylogenetic position and molecular chronology of a colonial green flagellate, Stephanosphaera pluvialis (Volvocales, Chlorophyceae), among unicellular algae. J Eukaryot Microbiol 63:340–348. https://doi.org/10.1111/jeu.12283
Nimmo IC, Barbrook AC, Lassadi I et al (2019) Genetic transformation of the dinoflagellate chloroplast. eLife 8:e45292. https://doi.org/10.7554/eLife.45292
Onishi M, Pringle JR (2016) Robust transgene expression from bicistronic mRNA in the green alga Chlamydomonas reinhardtii. G3 (Bethesda) 6:4115–4125. https://doi.org/10.1534/g3.116.033035
R Core Team (2013) R: A language and environment for statistical computing. R Foundation for Statistical Computing, Vienna
Suttangkakul A, Sirikhachornkit A, Juntawong P et al (2019) Evaluation of strategies for improving the transgene expression in an oleaginous microalga Scenedesmus acutus. BMC Biotechnol 19:4. https://doi.org/10.1186/s12896-018-0497-z
Van P, Jiang Mike, Gottardo R, Finak G (2018) ggcyto: Visualize Cytometry data with ggplot. Version 1.22.0. Bioconductor version: Release (3.14). URL https://bioconductor.org/packages/ggcyto/
Yamano T, Iguchi H, Fukuzawa H (2013) Rapid transformation of Chlamydomonas reinhardtii without cell-wall removal. J Biosci Bioeng 115:691–694. https://doi.org/10.1016/j.jbiosc.2012.12.020
Yang H, Genot B, Duhamel S et al (2022) Organismal and cellular interactions in vertebrate-alga symbioses. Biochem Soc Trans 50:609–620. https://doi.org/10.1042/BST20210153
Zhang R, Patena W, Armbruster U et al (2014) High-throughput genoty** of green algal mutants reveals random distribution of mutagenic insertion sites and endonucleolytic cleavage of transforming DNA. Plant Cell 26:1398–1409. https://doi.org/10.1105/tpc.114.124099
Acknowledgements
We thank Dr. Cory Bishop for sharing his unpublished draft genomes of the algae O. amblystomatis, Chlamydomonas nasuta, Chlamydomonas moewusii, and Chlamydomonas pseudogloegama. Comparative analyses of those genomes enabled promoter region selection.
Funding
This work was funded by the Gordon and Betty Moore Foundation grant GBMF5604 and by NSF OIA-1826734.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Competing interests
No competing interests declared.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
13199_2022_861_MOESM1_ESM.pdf
Supplementary file1 (PDF 131 KB) Figure S1: Promoter selection using alignments of genomic DNA and coding sequences of target genes from related species. Cnas/C_nasuta = Chlamydomonas nasuta; Cpsued/C_pseudo = Chlamydomonas pseudogloegama; Cmoe = Chlamydomonas moewusii; Oamb/O_ambyst = Oophila amblystomatis
13199_2022_861_MOESM3_ESM.png
Supplementary file3 (PNG 771 KB) Figure S3: Oophila amblystomatis cells are sensitive to paromomycin. Oophila amblystomatis cultures in AF6 media 7 days after treatment with 20 and 40 µg/mL paromomycin
13199_2022_861_MOESM4_ESM.png
Supplementary file4 (PNG 1737 KB) Figure S4: Fluorescence micrographs of a brightly fluorescent cell (A) from population pRPS4-i-GFP-APHVIII-A and from representative cells (B) from transformations with each plasmid. Green fluorescence is not easily visible by the naked eye, but can be bright occasionally (A), and on average cells from transformed lines have brighter green fluorescence than untransformed, wild-type cells, as shown by flow cytometry in Figure 5 of the main text
13199_2022_861_MOESM5_ESM.txt
Supplementary file5 (TXT 18 KB) Supplementary data: Plasmid sequences (.txt files) created for this study. Sequenced regions are in upper case
Rights and permissions
About this article
Cite this article
Genot, B., Burns, J.A. Transformation of the symbiotic alga Oophila amblystomatis: a new tool for animal-algae symbiosis studies. Symbiosis 87, 143–151 (2022). https://doi.org/10.1007/s13199-022-00861-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s13199-022-00861-0