Abstract
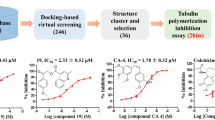
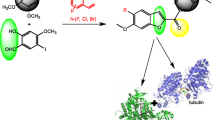
Virtual screening strategy was performed against the target β-tubulin to overcome paclitaxel resistance in blood cancer types. In essence, A185T and A248V are two such important mutations frequently observed in clinical trials that confer paclitaxel resistance. In the present investigation, compounds from NPACT database were filtered by pharmacokinetics, toxicity and binding energy values. A total of 5 active compounds were identified from a list of 1574 bioactive compounds investigated in our study. Finally, we have compiled all the characteristic features into biologically meaningful clusters by hierarchical clustering algorithm. Overall, the results from our analysis indicate that glaucarubol, isolated from the bark of Ailanthus excelsa tree, could be the potential lead molecule for the treatment of paclitaxel-resistant cancer types. It is worth stressing that our result is the first such observation of inhibitory action of glaucarubol against β-tubulin and warrants further experimental investigation.





Similar content being viewed by others
References
Mukhtar E, Adhami VM, Mukhtar H (2014) Targeting microtubules by natural agents for cancer therapy. Mol Cancer Ther 13:275–284
Gupta ML Jr, Bode CJ, Georg GI, Himes RH (2003) Understanding tubulin–Taxol interactions: mutations that impart Taxol binding to yeast tubulin. Proc Natl Acad Sci USA 100:6394–6397
Yin S, Bhattacharya R, Cabral F (2010) Human mutations that confer paclitaxel resistance. Mol Cancer Ther 9:327–335
Klebe G (2006) Virtual ligand screening: strategies, perspectives and limitations. Drug Discov Today 11:580–594
Kiang TK, Wilby KJ, Ensom MH (2015) A critical review on the clinical pharmacokinetics, pharmacodynamics, and clinical trials of ceftaroline. Clin Pharmacokinet 54:915–931
Khazir J, Bilal AM, Shabir AM, Don C (2013) Natural products as lead compounds in drug discovery. J Asian Nat Prod Res 15:764–788
Kingston DG (2011) Modern natural products drug discovery and its relevance to biodiversity conservation. J Nat Prod 74:496–511
Xu S, Chi S, ** Y, Shi Q, Ge M, Wang S et al (2012) Molecular dynamics simulation and density functional theory studies on the active pocket for the binding of paclitaxel to tubulin. J Mol Model 18:377–391
DeLano WL (2002) The PyMOL molecular graphics system. DeLano Scientific Alto, California
Mangal M, Sagar P, Singh H, Raghava GPS, Agarwal SM (2013) NPACT: naturally occurring plant-based anti-cancer compound-activity-target database. Nucleic Acids Res 41:1124–1129
Gasteiger J, Rudolph C, Sadowski J (1990) Automatic generation of 3D-atomic coordinates for organic molecules. Tetrahedron Comput Methodol Pergamon 3:537–547
Orr GA, Verdier-Pinard P, McDaid H, Horwitz SB (2003) Mechanisms of Taxol resistance related to microtubules. Oncogene 22:7280–7295
Guex N, Peitsch MC (1997) SWISS-MODEL and the Swiss-PdbViewer: an environment for comparative protein modeling. Electrophoresis 18:2714–2723
DE Pires V, Ascher DB, Blundell TL (2014) mCSM: predicting the effects of mutations in proteins using graph-based signatures. Bioinformatics 30:335–342
Raghav D, Sharma V (2013) An in silico evaluation of deleterious nonsynonymous single nucleotide polymorphisms in the ErbB3. Oncogene 2:206–211
Shoichet BK (2004) Virtual screening of chemical libraries. Nature 432:862–865
Bielska E, Lucas X, Czerwoniec A, Kasprzak JM, Kaminska KH, Bujnicki JM (2011) Virtual screening strategies in drug design—methods and applications. J Biotechnol Comput Biol Bionanotechnol 92:249–264
Tondi D, Slomczynska U, Costi MP, Watterson DM, Ghelli S, Shoichet BK (1999) Structure-based discovery and in-parallel optimization of novel competitive inhibitors of thymidylate synthase. Chem Biol 6:319–331
Merlot C (2010) Computational toxicology—a tool for early safety evaluation. Drug Discov Today 15:16–22
Balakin KV, Ivanenkov YA, Savchuk NP, Ivashchenko AA, Ekins S (2005) Comprehensive computational assessment of ADME properties using map** techniques. Curr Drug Discov Technol 2:99–113
Ertl P, Rohde B, Selzer P (2000) Fast calculation of molecular polar surface area as a sum of fragment-based contributions and its application to the prediction of drug transport properties. J Med Chem 43:3714–3717
Lipinski CA, Lombardo F, Dominy BW, Feeney PJ (1997) Experimental and computational approaches to estimate solubility and permeability in drug discovery and development settings. Adv Drug Deliv Rev 23:3–25
Lipinski CA (2004) Lead profiling lead- and drug-like compounds : the rule-of-five revolution. Drug Discov Today Technol 1:337–341
Muegge I (2003) Selection criteria for drug-like compounds. Med Res Rev 23:302–321
Buntrock RE (2002) ChemOffice Ultra 7.0. J Chem Inf Model 42:1505–1506
Tetko IV (2005) Computing chemistry on the web. Drug Discov Today 10:1497–1500
Sander T, Freyss J, von Korff M, Rufener C (2015) DataWarrior: an open-source program for chemistry aware data visualization and analysis. J Chem Inf Model 5:460–473
Von Korff M, Sander T (2006) Toxicity-indicating structural patterns. J Chem Inf Model 46:536–544
Olaniyan JM, Muhammad HL, Makun HA, Busari MB (2016) Acute and sub-acute toxicity studies of aqueous and methanol extracts of Nelsonia campestris in rats. J Acute Dis 5:62–70
Didziapetris R, Reynolds DP, Japertas P, Zmuidinavicius D, Petrauskas A (2006) In silico technology for identification of potentially toxic compounds in drug discovery. Curr Comput Aided Drug Des 2:95–103
Mazzatorta P, Estevez MD, Coulet M, Schilter B (2008) Modeling oral rat chronic toxicity. J Chem Inf Model 48:1949–1954
DE Pires V, Blundell TL, Ascher DB (2015) pkCSM: predicting small-molecule pharmacokinetic and toxicity properties using graph-based signatures. J Med Chem 58:4066–4072
Brown RD, Martin YC (1996) Use of structure–activity data to compare structure-based clustering methods and descriptors for use in compound selection. J Chem Inf Comput Sci 36:572–584
Pinheiro M, Afreixo V, Moura G, Freitas A, Santos MAS, Oliveira JL (2006) Statistical, computational and visualization methodologies to unveil gene primary structure features. Methods Inf Med 45:163–168
Perez F, Granger BE (2007) IPython: a system for interactive scientific computing. Comput Sci Eng 9:21–29
Akbari V, Moghim S, Reza Mofid M (2011) Comparison of epothilone and taxol binding in yeast tubulin using molecular modeling. Avicenna J Med Biotechnol 3:167–175
Natarajan K, Senapati S (2012) Understanding the basis of drug resistance of the mutants of αβ-tubulin dimer via molecular dynamics simulations. PLoS One 7:e42351
Ghanbarzadeh S, Ghasemi S, Shayanfar A, Ebrahimi-Najafabadi H (2015) 2D-QSAR study of some 2,5-diaminobenzophenone farnesyltransferase inhibitors by different chemometric methods. EXCLI J 14:484–495
Gopal V, Al Rashid MH, Majumder S, Maiti PP, Mandal SC (2015) Computational optimization of bioanalytical parameters for the evaluation of the toxicity of the phytomarker 1,4 naphthoquinone and its metabolite 1,2,4-trihydroxynapththalene. J Pharmacopunct 18:7–18
Morris GM, Goodsell DS, Halliday RS, Huey R, Hart WE, Belew RK et al (1998) Automated docking using a Lamarckian genetic algorithm and an empirical binding free energy function. J Comput Chem 19:1639–1662
Morris GM, Huey R, Lindstrom W, Sanner MF, Belew RK, Goodsell DS, Olson AJ (2009) AutoDock4 and AutoDockTools4: automated docking with selective receptor flexibility. J Comput Chem 30:2785–2791
Weiner SJ, Kollman PA, Case DA, Singh UC, Ghio C, Alagona G et al (1984) A new force field for molecular mechanical simulation of nucleic acids and proteins. J Am Chem Soc 106:765–784
Gasteiger J, Marsili M (1980) Iterative partial equalization of orbital electronegativity—a rapid access to atomic charges. Tetrahedron 36:3219–3228
Rathinasamy K, **dal B, Asthana J, Singh P, Balaji PV, Panda D (2010) Griseofulvin stabilizes microtubule dynamics, activates p53 and inhibits the proliferation of MCF-7 cells synergistically with vinblastine. BMC Cancer 10:213
Iman M, Davood A, Nematollahi AR, Dehpoor AR, Shafiee A (2011) Design and synthesis of new 1,4-dihydropyridines containing 4(5)-chloro-5(4)-imidazolyl substituent as a novel calcium channel blocker. Arch Pharm Res 34:1417–1426
Iman M, Saadabadi A, Davood A (2013) Docking studies of phthalimide pharmacophore as a sodium channel blocker. Iran J Basic Med Sci 16:1016–1021
Yu L, Pan Y, Wu Y (2009) Research on data normalization methods in multi-attribute evaluation. In: International conference on computational intelligence and software engineering (CiSE). IEEE, Wuhan, pp 1–5
Singhal S, Jeena M (2013) A study on WEKA tool for data preprocessing, classification and clustering. IJITEE 2:250–253
Hall M, Frank E, Holmes G, Pfahringer B, Reutemann P, Witten IH (2009) The WEKA data mining software. ACM 16(11):10
Wallace AC, Laskowski RA, Thornton JM (1995) LIGPLOT: a program to generate schematic diagrams of protein–ligand interactions. Protein Eng 8:127–134
Acknowledgements
The authors of the manuscript would like to thank the management of VIT University for providing the facility and support to carry out this research work.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Rights and permissions
About this article
Cite this article
Ramanathan, K., Verma, K., Gupta, N. et al. Discovery of Therapeutic Lead Molecule Against β-Tubulin Using Computational Approach. Interdiscip Sci Comput Life Sci 10, 734–747 (2018). https://doi.org/10.1007/s12539-017-0233-8
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12539-017-0233-8