Abstract
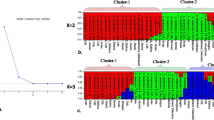
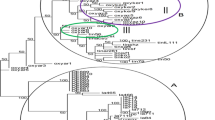
Saccharum spontaneum L. is the most diverse species of the genus Saccharum with the widest distribution from Africa-Mediterranean region to Far East including South Pacific islands. Its contribution in the development of present day sugarcane cultivars is well recognized. In the present study, 30 S. spontaneum L. accessions collected from North Western India including the states of Gujarat and Rajasthan were characterized with respect to their ecological distribution using 20 STMS (Sequence Tagged Microsatellite Site) primers. The study revealed the ability of STMS markers in discriminating the accessions with specific markers with 95.4% polymorphism. The pair wise genetic distance among 30 S. spontaneum L. accessions ranged from 0.17 to 0.48 with an average of 0.32. The maximum genetic distance was observed between the pairs IND 07-1462 and IND 08-1501 and IND 07-1464 and IND 08-1501. All the 30 S. spontaneum L. accessions were grouped into five major clusters. Among the five clusters, Cluster II had maximum of nine accessions viz., IND 07-1461, IND 07-1462, IND 07-1463, IND 07-1465, IND 07-1466, IND 07-1467, IND 07-1468, IND 07-1470 and IND 07-1474. All the accessions except IND 07-1474 were from Kachchh district of North West arid zone with low rainfall and high salinity. Majority of the accessions clustered as per the agro-climatic zones. Unique markers for the accession IND 07-1460, IND 07-1463 and IND 07-1482 were identified which could help in maintaining germplasm identity. Accessions from arid and semi-arid zones were grouped separately into different clusters. These accessions were collected from the regions where drought and salinity were prevalent hence could be the potential sources for develo** climate resilient sugarcane varieties with drought and salinity tolerance.



Similar content being viewed by others
References
Abraham, Z., R. Senthil Kumar, K. Joseph John, T.V.R.S. Sharma, N.V. Nair, M. Unnikrishnan, P.M. Kumaran, J.K. George, S. Uma, M. Latha, S.S. Malik, S.K. Mishra, D.C. Bhandari, and S.K. Pareek. 2008. Collection of Plant genetic resources from Andaman and Nicobar Islands. Genetic Resources and Crop Evolution 55: 1279–1289.
Aitken, K., J. Li, G. Piperidis, C. Qing, F. Yuanhong, and P. Jackson. 2018. Worldwide genetic diversity of the wild species Saccharum spontaneum L. and level of diversity captured within sugarcane breeding programs. Crop Breeding and Genetics 58: 218–229.
Ali, A., Y.B. Pan, Q.N. Wang, J.D. Wang, J.L. Chen, and S.J. Gao. 2019. Genetic diversity and population structure analysis of Saccharum and Erianthus genera using microsatellite (SSR) markers. Scientific Reports 9 (1): 1–10.
Ashkar, I.A., A. Alderfasi, W.B. Romdhane, M.F. Seleiman, R.A.E. Said, and A.A. Doss. 2020. Morphological and genetic diversity within salt tolerance detection in eighteen wheat genotypes. Plants 9 (3): 287.
Gouy, M., Y. Rousselle, A.T. Chane, A. Anglade, S. Royaert, S. Nibouche, and L. Costet. 2015. Genome wide association map** of agro-morphological and disease resistance traits in sugarcane. Euphytica 202: 269–284.
Govindaraj, P., R. Ramesh, C. Appunu, S. Swapna, and P.J. Priji. 2012. DNA Fingerprinting of sugarcane (Saccharum spp.) genotypes using sequence tagged microsatellite sites (STMS) markers. Plant Archieves 12: 347–352.
Govindaraj, P., R. Sindhu, A. Balamurugan, and C. Appunu. 2011. Molecular diversity in sugarcane hybrids (Saccharum spp complex) grown in Peninsular and East Coast zones of tropical India. Sugar Tech 13: 206–213.
Govindaraj, P., U.S. Natarajan, N. Balasundaram, M.N. Premachandran, T.R. Sharma, and N.K. Singh. 2005. Development of new microsatellite markers for the identification of interspecific hybrids in sugarcane. Sugarcane International 23: 30–34.
Govindaraj, P., S. Karthigeyan, and S.P. Adhini. 2016. Exploration and genetic diversity analysis of Saccharum spontaneum in Maharashtra State. Journal of Sugarcane Research 6 (2): 72–84.
Govindaraj, P., V.A. Amalraj, K. Mohanraj, and N.V. Nair. 2014. Collection, characterization and phenotypic diversity of Saccharum spontaneum L. from arid and semi-arid zones of Northwestern India. Sugar Tech 16: 36–43.
Jaccard, P. 1980. Nouvelles rescherches sur la distribution florale, Bulletin de la Societe Vaudoise des Sciences. Naturelles 44: 223–270.
Jiang, G.L. 2013. Molecular markers and marker-assisted breeding in plants. In: Plant breeding from laboratories to fields Editor, Andersen S.B. InTech Publishers p: 45–83
Karakoy, T., F.S. Baloch, F. Toklu, and H. Ozkan. 2014. Variation for selected morphological and quality-related traits among 178 faba bean landraces collected from Turkey. Plant Genetic Resources 12 (1): 5–13.
Meng, Z., J. Han, Y. Lin, Y. Zhao, Q. Lin, X. Ma, J. Wang, M. Zhang, L. Zhang, Q. Yang, and K. Wang. 2020. Characterization of a Saccharum spontaneum L. with a basic chromosome number of x = 10 provides new insights on genome evolution in genus Saccharum. Theoretical and Applied Genetics 133: 187–199.
Murray, M.G., and W.F. Thompson. 1980. Rapid isolation of high molecular weight plant DNA. Nucleic Acid Research 8: 4321–4325.
Nadeem, M.A., M.A. Nawaz, M.Q. Shahid, Y. Dogan, G. Comertpay, M. Yıldız, R. Hatipoglu, F. Ahmad, A. Alsaleh, N. Labhane, H. Ozkan, G. Chung, and F.S. Baloch. 2018. DNA molecular markers in plant breeding: Current status and recent advancements in genomic selection and genome editing. Biotechnology and Biotechnological Equipment. 32 (2): 261–285.
Nair, N.V., A.W. Jebadhas, T.V. Sreenivasan, and B.D. Sharma. 1991. Sugarcane germplasm collection in Manipur and Meghalaya. Indian Journal of Plant Genetic Resources 4 (1): 34–39.
Nair, N.V., A.W. Jebadhas, and T.V. Sreenivasan. 1993. Saccharum germplasm collection in Arunachal Pradesh. Indian Journal Plant Genetic Resources 6 (1): 21–26.
Nair, N.V., and S. Sekharan. 2009. Saccharum germplasm collection in Mizoram, India. Sugar Tech 11 (3): 288–291.
Nair, N.V., R. Nagarajan, and V.A. Amalraj. 2006. Saccharum Germplasm Collection from the Cauvery river basin and Coastal Tamil Nadu, India. IPGRI Plant Genetics Resource Newsletter 146: 56–59.
Pan, Y.B., M. Burner, B.L. Legendre, M.P. Grisham, and W.H. White. 2004. An assessment of the genetic diversity within a collection of Saccharum spontaneum L. L. with RAPD-PCR. Genetic Resources and Crop Evolution 51: 895–903.
Pan, Y.B., P. Liu, and Y. Que. 2015. Independently segregating simple sequence repeats (SSR) alleles in polyploid sugarcane. Sugar Tech 17 (3): 235–242.
Panje, R.R., and C.N. Babu. 1960. Studies of Saccharum spontaneum L. distribution and geographic association of chromosome numbers. Cytologia 25: 150–152.
Parthiban, S., P. Govindaraj, and S. Senthilkumar. 2018. Comparison of relative efficiency of genomic SSR and EST-SSR markers in estimating genetic diversity in sugarcane. 3 Biotech 8(3):144.
Pastina, M.M., M. Malosetti, R. Gazaffi, M. Mollinari, G.R.A. Margarido, K.M. Oliveira, L.R. Pinto, A.P. Souza, F.A. van Eeuwijk, and A.A.F. Garcia. 2012. A mixed model QTL analysis for sugarcane multiple-harvest-location trial data. Theoretical and Applied Genetics 124: 835–849.
Premachandran, M.N., S. Arvinth, and R. Lalitha. 2006. Chloroplast DNA polymorphism in psbC-trnS and trnL intron segments differentiate Saccharum and Erianthus. Indian Journal of Genetics 66: 283–286.
Rao, N. 2004. Plant genetic resources: advancing conservation and use through biotechnology. African Journal of Biotechnology 3: 136–145.
Rohlf, F. J. 1998. NTSYS-pc numerical taxonomy and multivariate analysis system, version 2.1. (Exeter Publ, New York).
Selvi, A., N.V. Nair, N. Balasundaram, and T. Mohapatra. 2003. Evaluation of maize microsatellite markers for genetic diversity analysis and fingerprinting in sugarcane. Genome 46 (3): 394–403.
Sheji, M., N.V. Nair, P.K. Chaturvedi, and A. Selvi. 2006. Analysis of Genetic Diversity among Saccharum spontaneum L. from Four geographical regions of India, using molecular arkers. Genetic Resources and Crop Evolution 53: 1221–1231.
Singh, R.K., N. Banerjee, M.S. Khan, S. Yadav, S. Kumar, S.K. Duttamajumder, R.J. Lal, J.D. Patel, H. Guo, D. Zhang, and A.H. Paterson. 2016. Identification of putative candidate genes for red rot resistance in sugarcane (Saccharum species hybrid) using LD-based association map**. Molecular Genetics and Genomics 291: 1363–1377.
Singh, R.K., S.K. Mishra, S.P. Singh, N. Mishra, and M.L. Sharma. 2010. Evaluation of microsatellite markers for genetic diversity analysis among sugarcane species and commercial hybrids. Australian Journal of crop Science 4: 115–124.
Singh, R.K., S.N. Jena, S. Khan, S. Yadav, N. Banarjee, S. Raghuvanshi, V. Bhardwaj, S.K. Dattamajumder, R. Kapur, S. Solomon, and M. Swapna. 2013. Development, cross-species/genera transferability of novel EST-SSR markers and their utility in revealing population structure and genetic diversity in sugarcane. Gene 524 (2): 309–329.
Singh, R.B., B. Singh, and R.K. Singh. 2019a. Cross-taxon transferability of sugarcane expressed sequence tags derived microsatellite (EST-SSR) markers across the related cereal grasses. Journal of Plant Biochemistry and Biotechnogy 28: 176–188.
Singh, R.B., B. Singh, and R.K. Singh. 2019b. Identification of elite Indian sugarcane varieties through DNA fingerprinting using genic microsatellite markers. Vegetos 32: 547–555.
Singh, R.K., R.B. Singh, S.P. Singh, and M.L. Sharma. 2011. Identification of sugarcane microsatellites associated to sugar content in sugarcane and transferability to other cereal genomes. Euphytica 182: 335–354.
Singh, R.K., S. Srivastava, S.P. Singh, M.L. Sharma, T. Mohopatra, N.K. Singh, and S.B. Singh. 2008. Identification of new microsatellite DNA markers for sugar and related traits in sugarcane. Sugar Tech 10: 327–333.
Siraree, A., N. Banerjee, S. Kumar, M.S. Khan, P.K. Singh, S. Sharma, R.K. Singh, and J. Sing. 2017. Identification of marker-trait associations for morphological descriptors and yield component traits in sugarcane. Physiology and Molecular Biology of Plants 23: 185–196.
Sundaram, S.K., M.S. Krishnamurthi, S. Rajeshwari, S. Sekar, and M. Shanmuganathan. 2010. Genetic base broadening of sugarcane (Saccharum spp.) by introgression of genes through intergeneric hybridization. Proceedings of the International Society of Sugar Cane Technologists 27: 1–9.
Tai, P.Y.P., and J.D. Miller. 2002. Germplasm diversity among four sugarcane species for sugar composition. Crop Science 42: 958–964.
Tai, P.Y.P., J.D. Miller, and B.L. Legendre. 1995. Evaluation of the world collection of Saccharum spontaneum L. Proceedings of International Society of Sugarcane Technologist 21: 250–260.
Todd, J.R., H. Sandhu, J. Binder, R. Arundale, V. Gordon, J. Sing, B. Glaz, and J. Wang. 2017. Fiber composition of a diversity panel of the world collection of sugarcane (Saccharum spp.) and related grasses. Bragantia 77: 175–192.
Tsuruta, S.I., M. Ebina, M. Kobayashi, W. Takahashi, and Y. Terajima. 2017. Development and validation of genomic simple sequence repeat markers in Erianthus arundinaceus. Molecular Breeding 37: 71.
Wang, Z., Y. Pan, J. Luo, Q. You, L. Xu, H. Zhang, and Y. Que. 2020. SSR-based genetic identity of sugarcane clones and its potential application in breeding and variety extension. Sugar Tech 22: 367–378.
Yu, X.H., X.H. Wang, and Q.H. Yang. 2019. Genetic diversity and phylogenetic relationship of Saccharum spontaneum L. with different ploidy levels based on SRAP markers. Sugar Tech 21: 802–814.
Zhang, G., Y. Li, W. He, X. Liu, H. Song, H. Liu, R. Zhu, and W. Fang. 2010. Analysis of the genetic diversity in Saccharum spontaneum L. accessions from Guangxi Province of China with RAPD-PCR. Sugar Tech 12: 31–35.
Zhang, J.S., Q. Zhang, L.T. Li, H.B. Tang, Q. Zhang, Y. Chen, J. Arrow, X.T. Zhang, A.Q. Wang, C.Y. Miao, and R. Ming. 2018. Recent polyploidization events in three Saccharum founding species. Plant Biotechnology Journal 17: 264–274.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Govindaraj, P., Gowri, R., Mohanraj, K. et al. SSR Marker Based Molecular Genetic Diversity Analysis Among Saccharum spontaneum (L.) Collected from North Western Region of India. Sugar Tech 23, 730–740 (2021). https://doi.org/10.1007/s12355-021-00956-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12355-021-00956-w