Abstract
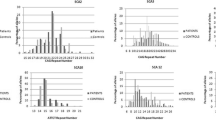
Spinocerebellar ataxia type 10 (SCA10) is a repeat expansion disease occurring mostly in Latin America, suggesting that the mutation spread with the peopling of the Americas, or that Amerindian populations, have a higher ATXN10 mutability. High frequency of large normal alleles is associated with prevalence and relative frequency of other repeat expansion diseases. To test whether the allele distribution of the SCA10-causing ATXN10 microsatellite in an Amerindian Peruvian population differs from that of other populations. The ATXN10 allele distribution in a Quechua Peruvian population from Puno, Peru, is similar to that of Finland. Mean allele size and mode were also similar to those of Mexico, Japan, and white Europeans. ATXN10 allele distribution in a healthy Amerindian population from Peru does not differ from that of other populations.


Similar content being viewed by others
References
Zu L, Figueroa KP, Grewal R, Pulst S-M. Map** of a new autosomal dominant spinocerebellar ataxia to chromosome 22. Am J Hum Genet. 1999;64:594–9.
Matsuura T, Yamagata T, Burgess DL, Rasmussen A, Grewal RP, Watase K, et al. Large expansion of the ATTCT pentanucleotide repeat in spinocerebellar ataxia type 10. Nat Genet. 2000;26:191–4.
Wang J, Wu Y, Lei L, Shen L, Jiang H, Zhou Y, et al. Polynucleotide repeat expansion of nine spinocerebellar ataxia subtypes and dentatorubral-pallidoluysian atrophy in healthy Chinese Han population. 2010.
Matsuura T, Fang P, Pearson CE, Jayakar P, Ashizawa T, Roa BB, et al. Interruptions in the expanded ATTCT repeat of spinocerebellar ataxia type 10: repeat purity as a disease modifier? Am J Hum Genet. 2006;78:125–9.
Alonso I, Jardim LB, Artigalas O, Saraiva-Pereira ML, Matsuura T, Ashizawa T, et al. Reduced penetrance of intermediate size alleles in spinocerebellar ataxia type 10. Neurology. 2006;66:1602–4.
Raskin S, Ashizawa T, Teive HA, Arruda WO, Fang P, Gao R, et al. Reduced penetrance in a Brazilian family with spinocerebellar ataxia type 10. Arch Neurol. 2007;64:591–4.
Matsuura T, Ashizawa T. Polymerase chain reaction amplification of expanded ATTCT repeat in spinocerebellar ataxia type 10. Ann Neurol. 2002;51:271–2.
Teive HAG, Munhoz RP, Raskin S, Arruda WO, de Paola L, Werneck LC, et al. Spinocerebellar ataxia type 10: frequency of epilepsy in a large sample of Brazilian patients. Mov Disord. 2010;25:2875–8.
Grewal RP, Achari M, Matsuura T, Durazo A, Tayag E, Zu L, et al. Clinical features and ATTCT repeat expansion in spinocerebellar ataxia type 10. Arch Neurol. 2002;59:1285–90.
Rasmussen A, Matsuura T, Ruano L, Yescas P, Ochoa A, Ashizawa T, et al. Clinical and genetic analysis of 4 Mexican families with spinocerebellar ataxia type 10. Ann Neurol. 2001;50:234–9.
Gatto EM, Gao R, White MC, Roca MCU, Etcheverry JL, Persi G, et al. Ethnic origin and extrapyramidal signs in an Argentinean spinocerebellar ataxia type 10 family. Neurology. 2007;69:216–8.
de Castilhos RM, Furtado GV, Gheno TC, Schaeffer P, Russo A, Barsottini O, et al. Spinocerebellar ataxias in Brazil—frequencies and modulating effects of related genes. Cerebellum. 2014;13:17–28.
Gheno TC, Furtado GV, Saute JAM, Donis KC, Fontanari AMV, Emmel VE, et al. Spinocerebellar ataxia type 10: common haplotype and disease progression rate in Peru and Brazil. Eur J Neurol. 2017;24:892–e36.
Abstracts of The Movement Disorder Society’s Thirteenth International Congress of Parkinson’s Disease and Movement Disorders. Mov Disord. 2009;24:S1–S653.
Roxburgh RH, Smith CO, Lim JG, Bachman DF, Byrd E, Bird TD. The unique co-occurrence of spinocerebellar ataxia type 10 (SCA10) and Huntington disease. J Neurol Sci. 2013;324:176–8.
Bampi GB, Bisso-Machado R, Hünemeier T, Gheno TC, Furtado GV, Veliz-Otani D, et al. Haplotype study in SCA10 families provides further evidence for a common ancestral origin of the mutation. NeuroMolecular Med. 2017;19:501–9.
Bushara K, Bower M, Liu J, McFarland KN, Landrian I, Hutter D, et al. Expansion of the spinocerebellar ataxia type 10 (SCA10) repeat in a patient with Sioux Native American ancestry. PLoS One. 2013;8:e81342.
Matsuura T, Ranum LPW, Volpini V, Pandolfo M, Sasaki H, Tashiro K, et al. Spinocerebellar ataxia type 10 is rare in populations other than Mexicans. Neurology. 2002;58:983–3.
Wang K, McFarland KN, Liu J, Zeng D, Landrian I, **a G, et al. Spinocerebellar ataxia type 10 in Chinese Han. Neurol Genet [Internet]. 2015 [cited 2018 Jan 10];1. Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC4809459/
Naito H, Takahashi T, Kamada M, Morino H, Yoshino H, Hattori N, et al. First report of a Japanese family with spinocerebellar ataxia type 10: the second report from Asia after a report from China. PLoS One. 2017;12:e0177955.
Takano H, Cancel G, Ikeuchi T, Lorenzetti D, Mawad R, Stevanin G, et al. Close associations between prevalences of dominantly inherited spinocerebellar ataxias with CAG-repeat expansions and frequencies of large normal CAG alleles in Japanese and Caucasian populations. Am J Hum Genet. 1998;63:1060–6.
Squitieri F, Andrew SE, Goldberg YP, Kremer B, Spence N, Zelsler J, et al. DNA haplotype analysis of Huntington disease reveals clues to the origins and mechanisms of CAG expansion and reasons for geographic variations of prevalence. Hum Mol Genet. 1994;3:2103–14.
Kay C, Collins JA, Wright GEB, Baine F, Miedzybrodzka Z, Aminkeng F, et al. The molecular epidemiology of Huntington disease is related to intermediate allele frequency and haplotype in the general population. Am J Med Genet B Neuropsychiatr Genet. 2018;177:346–57.
Semaka A, Kay C, Doty C, Collins JA, Bijlsma EK, Richards F, et al. CAG size-specific risk estimates for intermediate allele repeat instability in Huntington disease. J Med Genet. 2013;50:696–703.
Harris DN, Song W, Shetty AC, Levano KS, Cáceres O, Padilla C, et al. Evolutionary genomic dynamics of Peruvians before, during, and after the Inca Empire. PNAS. 2018;115:E6526–35.
Sandoval JR, Salazar-Granara A, Acosta O, Castillo-Herrera W, Fujita R, Pena SD, et al. Tracing the genomic ancestry of Peruvians reveals a major legacy of pre-Columbian ancestors. J Hum Genet. 2013;58:627–34.
Instituto Naional de Estadística e Informática. Resultados Definitivos de los Censos Nacionales 2017. Puno [Internet]. Lima, Perú: Instituto Nacional de Estadística e Informática; 2018 Oct. Available from: https://www.inei.gob.pe/media/MenuRecursivo/publicaciones_digitales/Est/Lib1563/.
Cornejo-Olivas M, Marca V, Dorschner MO, Inca Martinez M, Medina A, Shetty AC, et al. Target sequencing analysis of Parkinson’s disease genes in a healthy Amerindian population from Puno-Peru. San Diego, California, USA; 2014 [cited 2018 Jul 2]. Available from: http://www.ashg.org/2014meeting/abstracts/fulltext/f140122321.htm.
Cornejo-Olivas M, Torres L, Velit-Salazar MR, Inca-Martinez M, Mazzetti P, Cosentino C, et al. Variable frequency of LRRK2 variants in the Latin American research consortium on the genetics of Parkinson’s disease (LARGE-PD), a case of ancestry. NPJ Parkinsons Dis. 2017;3:19.
Arnold TB, Emerson JW, worldwide RCT and contributors. dgof: discrete goodness-of-fit tests [internet]. 2013 [cited 2018 Aug 2]. Available from: https://CRAN.R-project.org/package=dgof.
R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2013. 2014.
Raymond M, Rousset F. GENEPOP (version 1.2): population genetics software for exact tests and ecumenicism. J Hered. 1995;86:248–9.
Weir BS, Cockerham CC. Estimating F-statistics for the analysis of population structure. Evolution. 1984;38:1358–70.
Li H. Tabix: fast retrieval of sequence features from generic TAB-delimited files. Bioinformatics. 2011;27:718–9.
The 1000 Genomes Project Consortium. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65.
Juvonen V, Hietala M, Kairisto V, Savontaus M-L. The occurrence of dominant spinocerebellar ataxias among 251 Finnish ataxia patients and the role of predisposing large normal alleles in a genetically isolated population. Acta Neurol Scand. 2005;111:154–62.
Votsi C, Zamba-Papanicolaou E, Georghiou A, Kyriakides T, Papacostas S, Kleopa KA, et al. Investigation of SCA10 in the Cypriot population: further exclusion of SCA dynamic repeat mutations. J Neurol Sci. 2012;323:154–7.
Paradisi I, Ikonomu V, Arias S. Spinocerebellar ataxias in Venezuela: genetic epidemiology and their most likely ethnic descent. J Hum Genet. 2016;61:215–22.
Dakin EE, Avise JC. Microsatellite null alleles in parentage analysis. Heredity. 2004;93:504–9.
Almeida T, Alonso I, Martins S, Ramos EM, Azevedo L, Ohno K, et al. Ancestral origin of the ATTCT repeat expansion in spinocerebellar ataxia type 10 (SCA10). PLoS One. 2009;4:e4553.
Warby SC, Montpetit A, Hayden AR, Carroll JB, Butland SL, Visscher H, et al. CAG expansion in the Huntington disease gene is associated with a specific and targetable predisposing haplogroup. Am J Hum Genet. 2009;84:351–66.
Falush D, Almqvist EW, Brinkmann RR, Iwasa Y, Hayden MR. Measurement of mutational flow implies both a high new-mutation rate for Huntington disease and substantial underascertainment of late-onset cases. Am J Hum Genet. 2001;68:373–85.
Falush D. Haplotype background, repeat length evolution, and Huntington’s disease. Am J Hum Genet. 2009;85:939–42.
Li JZ, Absher DM, Tang H, Southwick AM, Casto AM, Ramachandran S, et al. Worldwide human relationships inferred from genome-wide patterns of variation. Science. 2008;319:1100–4.
Falush D, Iwasa Y. Size-dependent mutability and microsatellite constraints. Mol Biol Evol. 1999;16:960–6.
Templeton AR. Population genetics and microevolutionary theory. 1st ed. Hoboken, N.J: Wiley-Liss; 2006.
Matsuura T, Fang P, Lin X, Khajavi M, Tsuji K, Rasmussen A, et al. Somatic and germline instability of the ATTCT repeat in spinocerebellar ataxia type 10. Am J Hum Genet. 2004;74:1216–24.
Liu G, Bissler JJ, Sinden RR, Leffak M. Unstable spinocerebellar ataxia type 10 (ATTCT)·(AGAAT) repeats are associated with aberrant replication at the ATX10 locus and replication origin-dependent expansion at an ectopic site in human cells. Mol Cell Biol. 2007;27:7828–38.
Cherng N, Shishkin AA, Schlager LI, Tuck RH, Sloan L, Matera R, et al. Expansions, contractions, and fragility of the spinocerebellar ataxia type 10 pentanucleotide repeat in yeast. Proc Natl Acad Sci U S A. 2011;108:2843–8.
Walsh B, Lynch M. Evolution and selection of quantitative traits: I. Foundations. [Internet]. 1st ed. Unpublished manuscript [cited 2017 Mar 4]. Available from: nitro.biosci.arizona.edu/zbook/NewVolume_2/newvol2.html.
Acknowledgments
We are grateful to Victoria Marca and Karina Milla-Neyra for the administrative and logistic support as well as lab assistance.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
The sample donors gave informed consent for further use of their DNA samples in other genetic studies related to neurological disorders. The study was approved by the Institutional Review Board at Instituto Nacional de Ciencias Neurológicas (INCN).
Conflict of Interest
The authors declare that they have no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
About this article
Cite this article
Véliz-Otani, D., Inca-Martinez, M., Bampi, G.B. et al. ATXN10 Microsatellite Distribution in a Peruvian Amerindian Population. Cerebellum 18, 841–848 (2019). https://doi.org/10.1007/s12311-019-01057-x
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12311-019-01057-x