Abstract
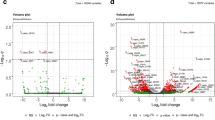
DNA methylation is the most important epigenetic modification involved in many essential biological processes. MET1 is one of DNA methyltransferases that affect the level of methylation in the entire genome. To explore the effect of MET1 gene silencing on gene expression profile of Chrysanthemum × morifolium 'Zi**gling'. The stem section and leaves at the young stage were taken for transcriptome sequencing. MET1-RNAi leaves had 8 differentially expressed genes while 156 differentially expressed genes were observed in MET1-RNAi stem compared with control leaves and stem. These genes encode many key proteins in plant biological processes, such as transcription factors, signal transduction mechanisms, secondary metabolite synthesis, transport and catabolism and interaction. In general, 34.58% of the differentially expressed genes in leaves and stems were affected by the reduction of the MET1 gene. The differentially expressed genes in stem and leaves of transgenic plants went through significant changes. We found adequate amount of candidate genes associated with flowering, however, the number of genes with significant differences between transgenic and control lines was not too high. Several flowering related genes were screened out for gene expression verification and all of them were obseved as consistent with transcriptome data. These candidate genes may play important role in flowering variation of chrysanthemum. This study reveals the mechanism of CmMET1 interference on the growth and development of chrysanthemum at the transcriptional level, which provides the basis for further research on the epigenetic regulation mechanism in flower induction and development.









Similar content being viewed by others
Availability and data materials
File contains detailed descriptions of all supplemental files. Transcriptomic data are available at NCBI with the accession number: SUB7902302. Gene sequences are available upon request. The authors affirm that all data necessary for confirming the conclusions of the article are present within the article, figures and tables.
References
Apweiler R, Bairoch A, Wu CH et al (2004) UniProt: the universal protein knowledgebase. Nucl Acids Res 32(90001):115–119. https://doi.org/10.1093/nar/gkh131
Arteaga-Vazquez M, Sidorenko L, Rabanal FA et al (2010) RNA-mediated trans-communcation can establish paramutation at the bllocus in maize. Proc Natl Acad Sci USA 107(29):12986–12991. https://doi.org/10.1073/pnas.1007972107
Ashburner M, Ball CA, Blake JA et al (2000) Gene ontology: tool for the unification of biology. Nat Genet Italic 25(1):25–29. https://doi.org/10.1038/75556
Baulcombe DC (2007) Molecular biology: Amplified silencing. Science 315(5809):199–200. https://doi.org/10.1126/science.1138030
Bewick AJ, Schmitz RJ (2017) Gene body DNA methylation in plants. Curr Opin Plant Biol 36:103–110. https://doi.org/10.1016/j.pbi.2016.12.007
Brettell RIS, Dennis ES (1991) Reactivation of a silent Ac following tissue culture is associated with heritable alterations in its methylation pattern. MGG-Mol Gen Genet 229(3):365–372. https://doi.org/10.1007/BF00267457
Burn JE, Bagnall DJ, Metzger JD et al (1993) DNA methylation, vernalization, and the initiation of flowering. Proc Natl Acad Sci USA 90(1):287–291. https://doi.org/10.1073/pnas.90.1.287
Cao XF, Jacobsen SE (2002) Locus-specific control of asymmetric and CpNpG methylation by the DRM and CMT3 methyltransferase genes. Proc Natl Acad Sci USA 99(4):16491–16498. https://doi.org/10.1073/pnas.162371599
Chen ZJ, Ni Z (2006) Mechanisms of genomic rearrangements and gene expression changes in plant polyploids. BioEssays 28(3):240–252. https://doi.org/10.1002/bies.20374
Cokus SJ, Feng S, Zhang X et al (2008) Shotgun bisulphite sequencing of the Arabidopsis genome reveals DNA methylation patterning. Nature 452(7184):215–219. https://doi.org/10.1038/nature06745
Deng YY, Li JQ, Wu SF et al (2006) Integrated NR database in protein annotation system and its localization. Comput Eng Ital 32(5):71–74. https://doi.org/10.1109/INFOCOM.2006.241
Dong W, Li M, Li Z, Li S, Zhu Y, Ding H, Wang Z (2020) Transcriptome analysis of the molecular mechanism of Chrysanthemum flower color change under short-day photoperiod. Plant Physiol Biochem 146:315–328. https://doi.org/10.1016/j.plaphy.2019.11.027
Eddy SR (1998) Profile hidden Markov models. Bioinformatics 14(9):755–763. https://doi.org/10.1198/016214502388618870
Eszter H, Szalai G, Janda T, Emil P, Ilona R, Demeter L (2003) Effect of vernalisation and 5-azacytidine on the methylation level of DNA in wheat (Triticum aestivum L. cv. martonvásár). Plant Sci 165(4):692. https://doi.org/10.1016/S0168-9452(03)00221-8
Fieldes MA (1994) Heritable effects of 5-azacytidine treatments on the growth and development of flax (Linum usitatissimum) genotrophs and genotypes. Genome 37(1):1–11. https://doi.org/10.1139/g94-001
Finn RD, Bateman A, Clements J et al (2014) Pfam: the protein families database. Nucl Acids Res 42:222–230. https://doi.org/10.1093/nar/gkt1223
Finnegan EJ, Genger RK, Kovac K et al (1998a) DNA methylation and the promotion of flowering by vernalization. Proc Natl Acad Sci USA 95(10):5824–5829
Finnegan EJ, Genger RK, Peacock WJ et al (1998b) DNA methylation in plants. Ann Rev Plant Physiol Plant Mol Biol 49(1):223–247
Fu XD, Kohli A, Twyman RM et al (2000) Alternative silencing effects involve distinct types of non-spreading cytosine methylation at a three-gene, single-copy transgenic locus in rice. MGG—Mol Gen Genet 263(1):106–118. https://doi.org/10.1007/PL00008669
Gao W, Li S, Li Z, Huang Y, Deng C, Lu L (2014) Detection of genome DNA methylation change in spinach induced by 5-azac. Mol Cell Probes. https://doi.org/10.1016/j.mcp.2014.02.002
Gong Z, Morales-Ruiz T, Ariza RR et al (2002) ROS1, a repressor of transcriptional gene silencing in Arabidopsis, encodes a DNA glycosylaselyase. Cell 111(6):803–814. https://doi.org/10.1016/S0092-8674(02)01133-9
Jaime HC, Damian S, Kristoffer F et al (2016) eggNOG 4.5: a hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucl Acids Res 44:286–293. https://doi.org/10.1093/nar/gkv1248
Kanehisa M, Goto S, Kawashima S et al (2004) The KEGG resource for deciphering the genome. Nucl Acids Res 32:277–280. https://doi.org/10.1093/nar/gkh063
Kang DR, Dai SL, Gao K, Zhang F, Luo H (2019a) Morphological variation of Chrysanthemum lavandulifolium induced by 5-azaC treatment. Sci Hortic 257:108645. https://doi.org/10.1016/j.scienta.2019.108645
Kang DR, Dai SL, Gao K, Zhang F, Luo H (2019) Morphological variation of Five Cut Chrysanthemum Cultivars Induced by 5-Azacytidine Treatment. Hortscience 54(7):1208–1216. https://doi.org/10.21273/HORTSCI14012-18
Ke W, Bai ZY, Liu QY et al (2018) Transcriptome analysis of chrysanthemum (Dendranthema grandiflorum) in response to low temperature stress. BMC Genom 19(1):319. https://doi.org/10.1186/s12864-018-4706-x
King GJ (1995) Morphological development in Brassica oleracea is modulated by in vivo treatment with 5-azacytidine. J Pomol Horticult Sci 70(2):333–342. https://doi.org/10.1080/14620316.1995.11515304
Kondo H, Shiraya T, Wada KC et al (2010) Induction of flowering by DNA demethylation in Perilla frutescens and Silene armeria: heritability of 5-azacytidine-induced effects and alteration of the DNA methylation state by photoperiodic conditions. Plant Sci 178(3):321–326. https://doi.org/10.1016/j.plantsci.2010.01.012
Koonin EV, Fedorova ND, Jackson JD et al (2004) A comprehensive evolutionary classification of proteins encoded in complete eukaryotic genomes. Genome Biol 5(2):1–28. https://doi.org/10.1186/gb-2004-5-2-r7
Lang Z, Wang Y, Tang K, Tang D, Datsenka T, Cheng J et al (2017) Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proc Natl Acad Sci. https://doi.org/10.1073/pnas.1705233114
Leng N, Dawson JA, Thomson JA et al (2013) EBSeq: an empirical Bayes hierarchical model for inference in RNA-seq experiments. Bioinformatics 29(8):1035–1043. https://doi.org/10.1093/bioinformatics/btt087
Li M, Guang Z, Zhu Z (2002) Effects of 5-azacytidine(5-azaC) and gibberellic acid(GA_3) on flowering of Brassica campestris ssp. Chinensis L. Master’s thesis, Journal of Shanghai Jiaotong University (in Chinese)
Li ZA, Li J, Liu YH et al (2016) DNA demethylation during chrysanthemum floral transition following short-day treatment. Electron J Biotechnol 21(3):77–81. https://doi.org/10.1016/j.ejbt.2016.02.006
Li SL, Li MM, Wang ZC et al (2019) Effects of the silencing of CmMET1 by RNA interference in chrysanthemum (Chrysanthemum morifolium). Plant Biotechnol Rep 13(1):63–72. https://doi.org/10.1007/s11816-019-00516-5
Li SF, Zhang GJ, Yuan JH et al (2015) Effect of 5-azaC on the growth, flowering time and sexual phenotype of spinach. Russ J Plant Physiol 62(5):670–675. https://doi.org/10.1134/s1021443715050118
Lipardi C, Wei Q, Paterson BM (2001) RNAi as random degradative PCR: siRNA primers convert mRNA into dsRNAs that are degraded to generate new siRNAs. Cell. https://doi.org/10.1016/s0092-8674(01)00537-2
Lister RO, Mallwey RC, Tonti-Filippini J et al (2008) Highly integrated single-base resolution maps of the epigenome in Arabidopsis. Cell 133(3):523–536. https://doi.org/10.1016/j.cell.2008.03.029
Livak KJ, Schmittgen TD (2002) Analysis of relative gene expression data using Real-Time quantitative PCR. Methods 75(4–5):402–408. https://doi.org/10.1006/meth.2001.1262
Lu J, Bi H, Zhang A et al (2018) Comparative transcriptome analysis by RNA-Seq of the regulation of low temperature responses in Dendranthema morifolium. Hortic Environ Biotechnol 59:383–395. https://doi.org/10.1007/s13580-018-0042-y
Lu G, Wu X, Chen B et al (2006) Detection of DNA methylation changes during seed germination in rapeseed (Brassica napus). Chin Sci Bull 51(2):182–190. https://doi.org/10.1007/s11434-005-1191-9
Matzke MA (2000) Transgene silencing by the host genome defense: implications for the evolution of epigenetic control mechanisms in plants and vertebrates. Plant Mol Biol 43(2/3):401–415. https://doi.org/10.1023/A:1006484806925
Messeguer R, Ganal MW, Steffens JC et al (1991) Characterization of the level, target sites and inheritance of cytosine methylation in tomato nuclear DNA. Plant Mol Biol 16(5):753–770. https://doi.org/10.1007/BF00015069
Nakano Y, Steward N, Sekine M et al (2000) A tobacco NtMET1 cDNA encoding a DNA methyltransferase: molecular characterization and abnormal phenotypes of transgenic tobacco plants. Plant Cell Physiol 1(4):448–457. https://doi.org/10.1093/pcp/41.4.448
Nie LJ, He YX, Wang ZC et al (2009) The effect of 5-azacytidine to the DNA methylation and morphogenesis character of Chrysanthemum during in vitro growth. Acta Horticult Sin 36:1783–1790 ((in Chinese))
Sako K, Maki Y, Kanai T, Kato E, Maekawa S et al (2012) Arabidopsis RPT2a 19S proteasome subunit, regulates gene silencing via DNA Methylation. PLoS One 7(5):e37086. https://doi.org/10.1371/journal.pone.0037086
Shafiq S, Berr A, Shen WH (2014) Combinatorial functions of diverse histone methylations in Arabidopsis thaliana flowering time regulation. New Phytol. https://doi.org/10.1111/nph.12493
Sheldon CC, Burn JE, Perez PP et al (1999) The FLF MADS box gene: a repressor of flowering in arabidopsis regulated by vernalization and methylation. Plant Cell 11(3):445–458. https://doi.org/10.1105/tpc.11.3.445
Smykal P, Valledor L, Rodríguez R (2007) Assessment of genetic and epigenetic stability in long-term in vitro shoot culture of pea (Pisum sativum L.). Plant Cell Reports. 26(11):1985–1998. https://doi.org/10.1007/s00299-007-0413-9
Tatusov RL, Galperin MY, Natale DA (2000) The COG database: a tool for genome scale analysis of protein functions and evolution. Nucl Acids Res 28(1):33–36. https://doi.org/10.1093/nar/28.1.33
Wang HB, Chen SM, Jiang JF et al (2015) Reference gene selection for cross-species and cross-ploidy level comparisons in Chrysanthemum spp. Sci Rep 5(1):8094. https://doi.org/10.1038/srep08094
Wang RY, Wang HG, Liu XY, Lian S, Chen L, Qiao ZJ, McInerney CE, Wang L (2017) Drought-induced transcription of resistant and sensitive common millet varieties. Anim Plant Sci 27(4):1303–1314
Wu CT, Morris JR (2001) Genes genetics and epigenetics: a correspondence. Science 293(5532):1103–1105. https://doi.org/10.1126/science.293.5532.1103
**ao W, Brown RC, Lemmon BE, Harada JJ, Fischer GRL (2006) Regulation of seed size by hypomethylation of maternal and paternal genomes. Plant Physiol 142(3):1160–1168. https://doi.org/10.1104/pp.106.088849
**e C, Mao X, Huang J, Ding Y et al (2011) KOBAS 2.0: a web server for annotation and identification of enriched pathways and diseases. Nucl Acids Res 39:316–322. https://doi.org/10.1093/nar/gkr483
**ong LZ, Xu CG, Maroof MAS et al (1999) Patterns of cytosine methylation in an elite rice hybrid and its parental lines, detected by a methylation-sensitive amplification polymorphism technique. Mol Gen Genet 261(3):439–446. https://doi.org/10.1007/s004380050986
Yang H, Han Z, Cao Y et al (2012) A companion cell-dominant and developmentally regulated H3K4 demethylase controls flowering time in arabidopsis via the repression of FLC expression. PLoS Genet 8(4):e1002664. https://doi.org/10.1371/journal.pgen.1002664
Zakrzewski F, Schmidt M, Van LM et al (2017) DNA methylation of retrotransposons, DNA transposons and genes in sugar beet (Beta vulgaris L.). Plant J Cell&Amp Mol Biol 90(6):1156–75. https://doi.org/10.1111/tpj.13526
Zhan J, Thakare D, Ma C et al (2015) RNA sequencing of laser-capture microdissected compartments of the maize kernel identifies regulatory modules associated with endosperm cell differentiation. Plant Cell 27(3):513–31. https://doi.org/10.1105/tpc.114.135657
Zhang X, Yazaki J, Sundaresan A et al (2006) Genome-wide high-resolution map** and functional analysis of DNA methylation in Arabidopsis. Cell 126(6):1189–1201. https://doi.org/10.1016/j.cell.2006.08.003
Zhao F, Zhang Q, Yan Y, Jia H, Han G (2019) Antioxidant constituents of chrysanthemum “**sidaju” cultivated in kaifeng. Fitoterapia. https://doi.org/10.1016/j.fitote.2019.02.003
Zhu QQ (2014) Effect of 5-azaC on DNA methylation and gene-expression of chrysanthemum. Master’s thesis, Henan University (in Chinese)
Acknowledgements
This research was funded by National Key Research and Development Program (2018YFD1000403), National Natural Science Foundation of China (31372090) and Science and Technology Development Project of Henan (182300410026).
Author information
Authors and Affiliations
Contributions
L.S.L\ZY performed experiments and helped in data analysis. LSL and ZY analyzed data. KDR wrote the manuscript with contributions from all authors. WZC guided the design of the whole test scheme. All authors have read and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Supplementary Materials: Table S1. Sequences for fluorescent quantitative. Table S2. Overview of the sequencing and assembly. Table S3. Length distribution of the transcripts and unigenes. Table S4. Functional annotation of unigenes. Table S5. Details of 122 different gene annotation results.
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Kang, Dr., Zhu, Y., Li, Sl. et al. Transcriptome analysis of differentially expressed genes in chrysanthemum MET1 RNA interference lines. Physiol Mol Biol Plants 27, 1455–1468 (2021). https://doi.org/10.1007/s12298-021-01022-1
Received:
Revised:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12298-021-01022-1