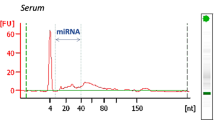
Abstract
Hemolysis has been known to affect the measurement of circulating biomarkers. In this study, clinically applicable procedures for microRNA (miRNA) detection in serum samples of acute myocardial infarction patients were established. The 89 samples from patients admitted to the coronary care unit were collected. These samples obtained from heparin-treated and untreated patients were subjected to heparinase digestion prior to miRNA measurements by multiplex RT-qPCR. The good reproducibility of miRNA detection after heparinase digestion (average R2 = 0.97) indicated that this method can be used routinely for samples regardless of heparin medication. Additionally, the degree of hemolysis in these samples was highly related to the hemoglobin absorbance at 414 nm. Based on the hemoglobin absorbance, five hemolysis-associated miRNAs were identified in our data normalized with respect to both the spike-in control and the RNA amount in a given sample. Using these calibration procedures, miRNAs can be accurately quantified and identified for clinical samples.

The practical procedures for miRNA detection in serum samples from the coronary care unit were established, and five hemolysis-associated miRNAs were accurately clarified through serial normalization.





Similar content being viewed by others
Abbreviations
- AMI:
-
acute myocardial infarction
- CCU:
-
coronary care unit
- conc.:
-
concentration
- CV:
-
coefficient of variation
- CVD:
-
cardiovascular disease
- miRNA:
-
microRNA
- RT-qPCR:
-
reverse transcription quantitative polymerase chain reaction
- SD:
-
standard deviation
- SE:
-
standard error
References
Ha, M., & Kim, V. N. (2014). Regulation of microRNA biogenesis. Nature Reviews. Molecular Cell Biology, 15(8), 509–524. https://doi.org/10.1038/nrm3838.
Mendell, J. T., & Olson, E. N. (2012). MicroRNAs in stress signaling and human disease. Cell, 148(6), 1172–1187. https://doi.org/10.1016/j.cell.2012.02.005.
Larrea, E., Sole, C., Manterola, L., Goicoechea, I., Armesto, M., Arestin, M., et al. (2016). New concepts in cancer biomarkers: circulating miRNAs in liquid biopsies. International Journal of Molecular Sciences, 17(5). https://doi.org/10.3390/ijms17050627.
Fan, P. C., Chen, C. C., Chen, Y. C., Chang, Y. S., & Chu, P. H. (2016). MicroRNAs in acute kidney injury. Human Genomics, 10(1), 29. https://doi.org/10.1186/s40246-016-0085-z.
Barwari, T., Joshi, A., & Mayr, M. (2016). MicroRNAs in cardiovascular disease. Journal of the American College of Cardiology, 68(23), 2577–2584. https://doi.org/10.1016/j.jacc.2016.09.945.
Viereck, J., & Thum, T. (2017). Circulating noncoding RNAs as biomarkers of cardiovascular disease and injury. Circulation Research, 120(2), 381–399. https://doi.org/10.1161/CIRCRESAHA.116.308434.
Boeckel, J. N., Thome, C. E., Leistner, D., Zeiher, A. M., Fichtlscherer, S., & Dimmeler, S. (2013). Heparin selectively affects the quantification of microRNAs in human blood samples. Clinical Chemistry, 59(7), 1125–1127. https://doi.org/10.1373/clinchem.2012.199505.
Kaudewitz, D., Lee, R., Willeit, P., McGregor, R., Markus, H. S., Kiechl, S., et al. (2013). Impact of intravenous heparin on quantification of circulating microRNAs in patients with coronary artery disease. Thrombosis and Haemostasis, 110(3), 609–615. https://doi.org/10.1160/TH13-05-0368.
Izraeli, S., Pfleiderer, C., & Lion, T. (1991). Detection of gene expression by PCR amplification of RNA derived from frozen heparinized whole blood. Nucleic Acids Research, 19(21), 6051. https://doi.org/10.1093/nar/19.21.6051.
Coelho-Lima, J., Mohammed, A., Cormack, S., Jones, S., Das, R., Egred, M., et al. (2018). Overcoming heparin-associated RT-qPCR inhibition and normalization issues for microRNA quantification in patients with acute myocardial infarction. Thrombosis and Haemostasis, 118(7), 1257–1269. https://doi.org/10.1055/s-0038-1660437.
Kondratov, K., Kurapeev, D., Popov, M., Sidorova, M., Minasian, S., Galagudza, M., et al. (2016). Heparinase treatment of heparin-contaminated plasma from coronary artery bypass grafting patients enables reliable quantification of microRNAs. Biomolecular Detectection and Quantification, 8, 9–14. https://doi.org/10.1016/j.bdq.2016.03.001.
Li, S., Zhang, F., Cui, Y., Wu, M., Lee, C., Song, J., et al. (2017). Modified high-throughput quantification of plasma microRNAs in heparinized patients with coronary artery disease using heparinase. Biochemical and Biophysical Research Communications, 493(1), 556–561. https://doi.org/10.1016/j.bbrc.2017.08.153.
Wu, C. S., Lin, F. C., Chen, S. J., Chen, Y. L., Chung, W. J., & Cheng, C. I. (2016). Optimized collection protocol for plasma microRNA measurement in patients with cardiovascular disease. BioMed Research International, 2016, 2901938. https://doi.org/10.1155/2016/2901938.
Kirschner, M. B., Edelman, J. J., Kao, S. C., Vallely, M. P., van Zandwijk, N., & Reid, G. (2013). The impact of hemolysis on cell-free microRNA biomarkers. Frontiers in Genetics, 4, 94. https://doi.org/10.3389/fgene.2013.00094.
Pritchard, C. C., Kroh, E., Wood, B., Arroyo, J. D., Dougherty, K. J., Miyaji, M. M., et al. (2012). Blood cell origin of circulating microRNAs: a cautionary note for cancer biomarker studies. Cancer Prevention Research (Philadelphia, Pa.), 5(3), 492–497. https://doi.org/10.1158/1940-6207.CAPR-11-0370.
Blondal, T., Jensby Nielsen, S., Baker, A., Andreasen, D., Mouritzen, P., Wrang Teilum, M., et al. (2013). Assessing sample and miRNA profile quality in serum and plasma or other biofluids. Methods, 59(1), S1–S6. https://doi.org/10.1016/j.ymeth.2012.09.015.
Shah, J. S., Soon, P. S., & Marsh, D. J. (2016). Comparison of methodologies to detect low levels of hemolysis in serum for accurate assessment of serum microRNAs. PLoS One, 11(4), e0153200. https://doi.org/10.1371/journal.pone.0153200.
Kroh, E. M., Parkin, R. K., Mitchell, P. S., & Tewari, M. (2010). Analysis of circulating microRNA biomarkers in plasma and serum using quantitative reverse transcription-PCR (qRT-PCR). Methods, 50(4), 298–301. https://doi.org/10.1016/j.ymeth.2010.01.032.
Pritchard, C. C., Cheng, H. H., & Tewari, M. (2012). MicroRNA profiling: approaches and considerations. Nature Reviews. Genetics, 13(5), 358–369. https://doi.org/10.1038/nrg3198.
Marabita, F., de Candia, P., Torri, A., Tegner, J., Abrignani, S., & Rossi, R. L. (2016). Normalization of circulating microRNA expression data obtained by quantitative real-time RT-PCR. Briefings in Bioinformatics, 17(2), 204–212. https://doi.org/10.1093/bib/bbv056.
Schwarzenbach, H., da Silva, A. M., Calin, G., & Pantel, K. (2015). Data normalization strategies for microRNA quantification. Clinical Chemistry, 61(11), 1333–1342. https://doi.org/10.1373/clinchem.2015.239459.
Wylie, D., Shelton, J., Choudhary, A., & Adai, A. T. (2011). A novel mean-centering method for normalizing microRNA expression from high-throughput RT-qPCR data. BMC Research Notes, 4, 555. https://doi.org/10.1186/1756-0500-4-555.
Vila-Navarro, E., Duran-Sanchon, S., Vila-Casadesus, M., Moreira, L., Gines, A., Cuatrecasas, M., et al. (2019). Novel circulating miRNA signatures for early detection of pancreatic neoplasia. Clinical and Translational Gastroenterology, 10(4), e00029. https://doi.org/10.14309/ctg.0000000000000029.
Xue, S., Liu, D., Zhu, W., Su, Z., Zhang, L., Zhou, C., et al. (2019). Circulating miR-17-5p, miR-126-5p and miR-145-3p are novel biomarkers for diagnosis of acute myocardial infarction. Frontiers in Physiology, 10, 123. https://doi.org/10.3389/fphys.2019.00123.
Thygesen, K., Alpert, J. S., White, H. D., & Joint ESC/ACCF/AHA/WHF Task Force for the Redefinition of Myocardial Infarction. (2007). Universal definition of myocardial infarction. Journal of the American College of Cardiology, 50(22), 2173–2195. https://doi.org/10.1016/j.jacc.2007.09.011.
Chang, P. Y., Chen, C. C., Chang, Y. S., Tsai, W. S., You, J. F., Lin, G. P., et al. (2016). MicroRNA-223 and microRNA-92a in stool and plasma samples act as complementary biomarkers to increase colorectal cancer detection. Oncotarget, 7(9), 10663–10675. https://doi.org/10.18632/oncotarget.7119.
Fan, P. C., Chen, C. C., Peng, C. C., Chang, C. H., Yang, C. H., Yang, C., et al. (2019). A circulating miRNA signature for early diagnosis of acute kidney injury following acute myocardial infarction. Journal of Translational Medicine, 17(1), 139. https://doi.org/10.1186/s12967-019-1890-7.
Marzi, M. J., Montani, F., Carletti, R. M., Dezi, F., Dama, E., Bonizzi, G., et al. (2016). Optimization and standardization of circulating microRNA detection for clinical application: the miR-test case. Clinical Chemistry, 62(5), 743–754. https://doi.org/10.1373/clinchem.2015.251942.
Mao, J., Lv, Z., & Zhuang, Y. (2014). MicroRNA-23a is involved in tumor necrosis factor-alpha induced apoptosis in mesenchymal stem cells and myocardial infarction. Experimental and Molecular Pathology, 97(1), 23–30. https://doi.org/10.1016/j.yexmp.2013.11.005.
Wang, Y., Pan, X., Fan, Y., Hu, X., Liu, X., **ang, M., et al. (2015). Dysregulated expression of microRNAs and mRNAs in myocardial infarction. American Journal of Translational Research, 7(11), 2291–2304.
Aranda, R. t., Dineen, S. M., Craig, R. L., Guerrieri, R. A., & Robertson, J. M. (2009). Comparison and evaluation of RNA quantification methods using viral, prokaryotic, and eukaryotic RNA over a 10(4) concentration range. Analytical Biochemistry, 387(1), 122–127. https://doi.org/10.1016/j.ab.2009.01.003.
Bae, I. S., Chung, K. Y., Yi, J., Kim, T. I., Choi, H. S., Cho, Y. M., et al. (2015). Identification of reference genes for relative quantification of circulating microRNAs in bovine serum. PLoS One, 10(3), e0122554. https://doi.org/10.1371/journal.pone.0122554.
Occhipinti, G., Giulietti, M., Principato, G., & Piva, F. (2016). The choice of endogenous controls in exosomal microRNA assessments from biofluids. Tumour Biology, 37(9), 11657–11665. https://doi.org/10.1007/s13277-016-5164-1.
Funding
This work was supported by the Chang Gung Medical Foundation (CIRPG3B0043, CLRPD1J0011, and CMRPG5G0111) and the Ministry of Science and Technology of Taiwan (106-2314-B-182A-118-MY3 and 107-2320-B-182-024). The funding organization(s) played no role in the study design; in the collection, analysis, and interpretation of data; in the writing of the report; or in the decision to submit the report for publication.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Material preparation, data collection, and analysis were performed by Chia-Chun Chen and Chen-Ching Peng. The sample collection and clinical data processing were performed by Pei-Chun Fan and Chih-Hsiang Chang. All authors commented on previous versions of the manuscript and approved the final manuscript.
Corresponding author
Ethics declarations
Conflict of Interest
The authors declare that they have no conflicts of interest.
Ethical Approval and Consent to Participate
The study protocol was approved by the Institutional Review Board of Chang Gung Medical Foundation, Taipei, Taiwan. The written informed consents were obtained from the patients themselves or legal representatives.
Animal Studies
No animal studies were carried out by the authors for this article.
Additional information
Associate Editor Yihua Bei oversaw the review of this article
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic Supplementary Material
ESM 1
(DOCX 4730 kb).
Rights and permissions
About this article
Cite this article
Chen, CC., Peng, CC., Fan, PC. et al. Practical Procedures for Improving Detection of Circulating miRNAs in Cardiovascular Diseases. J. of Cardiovasc. Trans. Res. 13, 977–987 (2020). https://doi.org/10.1007/s12265-020-10019-2
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12265-020-10019-2