Abstract
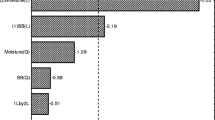
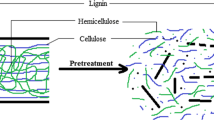
Cassava pulp (CP) is a starchy lignocellulose waste with starch granules entrapped within the cell wall. The enriched hydrolytic and acidogenic bacterial consortium (EHA) was established to break down the lignocellulosic structure and free starch granules during CP conversion into volatile fatty acids (VFAs). Consecutive batch subcultures were used to acclimatize the anaerobic lignocellulolytic microbial consortium for the anaerobic digestion of CP. Hydrolytic and acidogenic microorganisms in the anaerobic lignocellulolytic microbial consortium were then enriched by inhibiting methanogenic activity until a stable consortium, namely EHA4, was obtained. The performance of EHA4 in CP utilization was investigated based on different operating conditions, including CP concentration, inoculum-to-substrate ratio, and pH adjustment. The alpha diversity revealed that the acclimatization and enrichment decreased the consortium’s species richness, creating a solid community specializing in CP hydrolysis and acidification. EHA4 consortium used 100% of CP added within 4 days while producing high-yield VFAs dominated by butyrate and acetate. Clostridium was the dominant bacteria related to hydrolysis and acidification of CP found in EHA4 (41% relative abundance). The performance of EHA4 to degrade higher CP concentration showed a decrease in CP removal due to total VFAs inhibition while maintaining the pH recovered the CP utilization up to 4%. EHA4 is a potential microbial consortium for starchy lignocellulosic CP hydrolysis and acidification as the inoculum starter in biogas production in the future.





Similar content being viewed by others
Data availability
The datasets supporting this article’s conclusion are included in the article and its supplementary information files.
References
Sowcharoensuk C (2021) Industry outlook 2022-2024: cassava industry. Krungsri Research https://www.krungsri.com/en/research/industry/industry-outlook/agriculture/cassava/io/io-cassava-21. Accessed 26 October 2022
Lerdlattaporn R, Phalakornkule C, Trakulvichean S, Songkasiri W (2021) Implementing circular economy concept by converting cassava pulp and wastewater to biogas for sustainable production in starch industry. Sustain Environ Res 31(1):20. https://doi.org/10.1186/s42834-021-00093-9
Varongchayakul S, Songkasiri W, Chaiprasert P (2021) Optimization of cassava pulp pretreatment by liquid hot water for biomethane production. Bioenergy Res 14(4):1312–1327. https://doi.org/10.1007/s12155-020-10238-0
Marius Bredon JD, Noël C, Moumen B, Bouchon D (2018) Lignocellulose degradation at the holobiont level: teamwork in a keystone soil invertebrate. Microbiome. https://doi.org/10.1186/s40168-018-0536-y
Passanun Lomwongsopon NA (2022) Mild chemical pretreatment of cassava pulp for enhancing high-load anaerobic digestion. Bioresour Technol. https://doi.org/10.1016/j.biteb.2021.100896
Thongbunrod N, Chaiprasert P (2022) Potential of enriched and stabilized anaerobic lignocellulolytic fungi co-existing with bacteria and methanogens for enhanced methane production from rice straw. Biomass Convers Biorefin. https://doi.org/10.1007/s13399-022-03129-1
Kleerebezem R, van Loosdrecht MCM (2007) Mixed culture biotechnology for bioenergy production. Curr Opin Biotechnol 18(3):207–212. https://doi.org/10.1016/j.copbio.2007.05.001
Pason P, Tachaapaikoon C, Panichnumsin P, Ketbot P, Waeonukul R, Kosugi A et al (2020) One-step biohydrogen production from cassava pulp using novel enrichment of anaerobic thermophilic bacteria community. Biocatal Agric Biotechnol 27:101658. https://doi.org/10.1016/j.bcab.2020.101658
Xu Z, Jiang L (2011) 3.20 - Butyric Acid. In: Moo-Young M (ed) Comprehensive biotechnology, 2nd edn. Academic Press, Burlington
Tomazetto G, Hahnke S, Wibberg D, Pühler A, Klocke M, Schlüter A (2018) Proteiniphilum saccharofermentans str. M3/6T isolated from a laboratory biogas reactor is versatile in polysaccharide and oligopeptide utilization as deduced from genome-based metabolic reconstructions. Biotechnol Rep 18:e00254. https://doi.org/10.1016/j.btre.2018.e00254
Silva Rabelo CAB, Okino CH, Sakamoto IK, Varesche MBA (2020) Isolation of Paraclostridium CR4 from sugarcane bagasse and its evaluation in the bioconversion of lignocellulosic feedstock into hydrogen by monitoring cellulase gene expression. Sci Total Environ 715:136868. https://doi.org/10.1016/j.scitotenv.2020.136868
Hahnke S, Langer T, Koeck DE, Klocke M (2016) Description of Proteiniphilum saccharofermentans sp. nov., Petrimonas mucosa sp. nov. and Fermentimonas caenicola gen. nov., sp. nov., isolated from mesophilic laboratory-scale biogas reactors, and emended description of the genus Proteiniphilum. Int J Syst Evol Microbiol 66(3):1466–1475. https://doi.org/10.1099/ijsem.0.000902
Thongbunrod N, Chaiprasert P (2021) Efficacy and metagenomic analysis of the stabilized anaerobic lignocellulolytic microbial consortium from Bubalus bubalis rumen with rice straw enrichment for methane production. Bioenergy Res 14(3):870–890. https://doi.org/10.1007/s12155-020-10167-y
Ferraro A, Dottorini G, Massini G, Mazzurco Miritana V, Signorini A, Lembo G et al (2018) Combined bioaugmentation with anaerobic ruminal fungi and fermentative bacteria to enhance biogas production from wheat straw and mushroom spent straw. Bioresour Technol 260:364–373. https://doi.org/10.1016/j.biortech.2018.03.128
Basak B, Patil SM, Kumar R, Ahn Y, Ha G-S, Park Y-K et al (2022) Syntrophic bacteria- and Methanosarcina-rich acclimatized microbiota with better carbohydrate metabolism enhances biomethanation of fractionated lignocellulosic biocomponents. Bioresour Technol 360:127602. https://doi.org/10.1016/j.biortech.2022.127602
Sluiter A, Hames B, Hyman D, Payne C, Ruiz R, Scalata C, Sluiter J, Templeton D, Wolfe J (2008) Determination of total solids in biomass and total dissolved solids in liquid process samples. National Renewable Energy Laboratory http://purl.access.gpo.gov/GPO/LPS94120. Accessed 26 October 2022
McCleary BV, Gibson TS, Mugford DC et al (2020) Measurement of total starch in cereal products by Amyloglucosidase-α-Amylase Method: Collaborative Study. J AOAC Int 80(3):571–579. https://doi.org/10.1093/jaoac/80.3.571
Van Soest PJ, Robertson JB, Lewis BA (1991) Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74(10):3583–3597. https://doi.org/10.3168/jds.S0022-0302(91)78551-2
Cheng YF, Edwards JE, Allison GG, Zhu W-Y, Theodorou MK (2009) Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour Technol 100(20):4821–4828. https://doi.org/10.1016/j.biortech.2009.04.031
Ranjard L, Poly F, Lata J-C, Mougel C, Thioulouse J, Nazaret S (2001) Characterization of bacterial and fungal soil communities by automated ribosomal intergenic spacer analysis fingerprints: Biological and methodological variability. Appl Environ Microbiol 67(10):4479–4487. https://doi.org/10.1128/AEM.67.10.4479-4487.2001
Ciesielski S, Bułkowska K, Dabrowska D, Kaczmarczyk D, Kowal P, Możejko J (2013) Ribosomal intergenic spacer analysis as a tool for monitoring methanogenic Archaea changes in an anaerobic digester. Curr Microbiol 67(2):240–248. https://doi.org/10.1007/s00284-013-0353-2
Bolyen E, Rideout JR, Dillon MR, Bokulich NA, Abnet CC, Al-Ghalith GA et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37(8):852–857. https://doi.org/10.1038/s41587-019-0209-9
Beall SS, Jenkins D, Vidanage SA (1998) A systematic analytical artifact that significantly influences anaerobic digestion efficiency measurement. Water Environ Res 70(5):1019–1024
Panichnumsin P, Nopharatana A, Ahring BK, Chaiprasert P (2012) Enhanced biomethanation in co-digestion of cassava pulp and pig manure using a two-phase anaerobic system. J Renew Sust Energ 3:73–79
Park S-G, Rhee C, Shin SG, Shin J, Mohamed HO, Choi Y-J et al (2019) Methanogenesis stimulation and inhibition for the production of different target electrobiofuels in microbial electrolysis cells through an on-demand control strategy using the coenzyme M and 2-bromoethanesulfonate. Environ Int 131:105006. https://doi.org/10.1016/j.envint.2019.105006
Puengrang P, Suraraksa B, Prommeenate P, Boonapatcharoen N, Cheevadhanarak S, Tanticharoen M et al (2020) Diverse microbial community profiles of propionate-degrading cultures derived from different sludge sources of anaerobic wastewater treatment plants. Microorganisms 8(2):277. https://doi.org/10.3390/microorganisms8020277
Eryildiz B, Lukitawesa TMJ (2020) Effect of pH, substrate loading, oxygen, and methanogens inhibitors on volatile fatty acid (VFA) production from citrus waste by anaerobic digestion. Bioresour Technol 302:122800. https://doi.org/10.1016/j.biortech.2020.122800
Han Y, Green H, Tao W (2020) Reversibility of propionic acid inhibition to anaerobic digestion: Inhibition kinetics and microbial mechanism. Chemosphere 255:126840. https://doi.org/10.1016/j.chemosphere.2020.126840
Andermann T, Antonelli A, Barrett RL, Silvestro D (2022) Estimating alpha, beta, and gamma diversity through deep learning. Front Plant Sci 13:839407. https://doi.org/10.3389/fpls.2022.839407
De Vrieze J, Christiaens MER, Walraedt D, Devooght A, Ijaz UZ, Boon N (2017) Microbial community redundancy in anaerobic digestion drives process recovery after salinity exposure. Water Res 111:109–117. https://doi.org/10.1016/j.watres.2016.12.042
Saloua Biyada MM, Biyada S, Merzouki M, Dėmčėnko T, Vasiliauskienė D, Marčiulaitienė E, Vasarevičius S, Urbonavičius J (2022) The effect of feedstock concentration on the microbial community dynamics during textile waste composting. Front Ecol Evol 10(813488). https://doi.org/10.3389/fevo.2022.813488
Lagoa-Costa B, Kennes C, Veiga MC (2022) Influence of feedstock mix ratio on microbial dynamics during acidogenic fermentation for polyhydroxyalkanoates production. J Environ Manage 303:114132. https://doi.org/10.1016/j.jenvman.2021.114132
Hamdi O, Ben Hania W, Postec A, Bouallagui H, Hamdi M, Bonin P et al (2015) Aminobacterium thunnarium sp. nov., a mesophilic, amino acid-degrading bacterium isolated from an anaerobic sludge digester, pertaining to the phylum Synergistetes. Int J Syst Evol Microbiol 65(2):609–614. https://doi.org/10.1099/ijs.0.068965-0
Hernandez-Eugenio G, Fardeau M-L, Cayol J-L, Patel BKC, Thomas P, Macarie H et al (2002) Sporanaerobacter acetigenes gen. nov., sp. nov., a novel acetogenic, facultatively sulfur-reducing bacterium. Int J Syst Evol Microbiol 52(4):1217–1223. https://doi.org/10.1099/00207713-52-4-1217
Esquivel-Elizondo S, Bağcı C, Temovska M, Jeon BS, Bessarab I, Williams RBH et al (2020) The isolate Caproiciproducens sp. 7D4C2 produces n-caproate at mildly acidic conditions from hexoses: genome and rBOX comparison with related strains and chain-elongating bacteria. Front Microbiol 11(594524). https://doi.org/10.3389/fmicb.2020.594524
Laverde Gomez JA, Mukhopadhya I, Duncan SH, Louis P, Shaw S, Collie-Duguid E et al (2019) Formate cross-feeding and cooperative metabolic interactions revealed by transcriptomics in co-cultures of acetogenic and amylolytic human colonic bacteria. Environ Microbiol 21(1):259–271. https://doi.org/10.1111/1462-2920.14454
Duan Y, Zhou A, Yue X, Zhang Z, Gao Y, Luo Y et al (2020) Acceleration of the particulate organic matter hydrolysis by start-up stage recovery and its original microbial mechanism. Front Environ Sci Eng 15(1):12. https://doi.org/10.1007/s11783-020-1304-3
Iino T, Mori K, Tanaka K, Suzuki KI, Harayama S (2007) Oscillibacter valericigenes gen. nov., sp. nov., a valerate-producing anaerobic bacterium isolated from the alimentary canal of a Japanese corbicula clam. Int J Syst Evol Microbiol 57(8):1840–1845. https://doi.org/10.1099/ijs.0.64717-0
Lee GH, Rhee MS, Chang DH, Lee J, Kim S, Yoon MH et al (2013) Oscillibacter ruminantium sp. nov., isolated from the rumen of Korean native cattle. Int J Syst Evol Microbiol 63(6):1942–1946. https://doi.org/10.1099/ijs.0.041749-0
Pokusaeva K, Fitzgerald GF, van Sinderen D (2011) Carbohydrate metabolism in Bifidobacteria. Genes Nutr 6(3):285–306. https://doi.org/10.1007/s12263-010-0206-6
Kraatz M, Wallace RJ, Svensson L (2011) Olsenella umbonata sp. nov., a microaerotolerant anaerobic lactic acid bacterium from the sheep rumen and pig jejunum, and emended descriptions of Olsenella, Olsenella uli and Olsenella profusa. Int J Syst Evol Microbiol 61(4):795–803. https://doi.org/10.1099/ijs.0.022954-0
Smith MR, Mah RA (1981) 2-Bromoethanesulfonate: A selective agent for isolating resistant Methanosarcina mutants. Curr Microbiol 6(5):321–326. https://doi.org/10.1007/BF01566885
Basak B, Ahn Y, Kumar R, Hwang J-H, Kim K-H, Jeon B-H (2022) Lignocellulolytic microbiomes for augmenting lignocellulose degradation in anaerobic digestion. Trends Microbiol 30(1):6–9. https://doi.org/10.1016/j.tim.2021.09.006
Frassoldati A, Ranzi E (2019) Modeling of thermochemical conversion of biomasses. Reference module in chemistry, Molecular sciences and chemical engineering. Elsevier, Amsterdam
Zhang D, Wang Y, Zhang C, Zheng D, Guo P, Cui Z (2018) Characterization of a thermophilic lignocellulose-Degrading Microbial Consortium with High Extracellular Xylanase Activity. J Microbiol Biotechnol 28(2):305–313. https://doi.org/10.4014/jmb.1709.09036
Wambugu CW, Rene ER, Van de Vossenberg J, Dupont C, van Hullebusch ED (2020) Chapter 8 - Biochar from various lignocellulosic biomass wastes as an additive in biogas production from food waste. In: Bhaskar T, Pandey A, Rene ER, Tsang DCW (eds) Waste Biorefinery. Elsevier, Amsterdam
Ren J, Yuan X, Li J, Ma X, Zhao Y, Zhu W et al (2014) Performance and microbial community dynamics in a two-phase anaerobic co-digestion system using cassava dregs and pig manure. Bioresour Technol 155:342–351. https://doi.org/10.1016/j.biortech.2013.12.120
Darwin C-RR, Charles W (2018) Ethanol and lactic acid production from sugar and starch wastes by anaerobic acidification. Eng Life Sci 18(9):635–642. https://doi.org/10.1002/elsc.201700178
Masset J, Calusinska M, Hamilton C, Hiligsmann S, Joris B, Wilmotte A et al (2012) Fermentative hydrogen production from glucose and starch using pure strains and artificial co-cultures of Clostridium spp. Biotechnol Biofuels 5(1):35. https://doi.org/10.1186/1754-6834-5-35
Wang S, Xu C, Song L, Zhang J (2022) Anaerobic digestion of food waste and its microbial consortia: A historical review and future perspectives. Int J Environ Res Public Health 19(15). https://doi.org/10.3390/ijerph19159519
Acknowledgements
The authors thank Petchra Pra Jom Klao Ph.D. Research Scholarship (KMUTT-NSTDA) from King Mongkut’s University of Technology Thonburi and the National Science and Technology Development Agency for the financial support to Ms. Alifia Issabella Mulyawati and ECoWaste Laboratory for the laboratory facility.
Funding
This present research was financially supported by Petchra Pra Jom Klao Ph.D. Research Scholarship (KMUTT-NSTDA) from King Mongkut’s University of Technology Thonburi and National Science and Technology Development Agency (No. of agreement 3/2562). Research funding was also supported by Thailand Science Research and Innovation (TSRI), Ministry of Higher Education, Science, Research, and Innovation (MHESI) for Basic Research Fund (Fundamental Fund, FF 2566).
Author information
Authors and Affiliations
Contributions
Conceptualization, laboratory experiment, data curation, visualization, writing—original draft: Alifia Issabella Mulyawati; Conceptualization, validation, co-supervision, writing—review: Benjaphon Suraraksa; Conceptualization, methodology, validation, research supervision, resources, writing-review, and editing: Pawinee Chaiprasert.
Corresponding author
Ethics declarations
Consent to participate
All the listed authors agree.
Consent for publication
All the listed authors mutually agree and hope this original work can be published.
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
ESM 1
(DOCX 452 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mulyawati, A.I., Suraraksa, B. & Chaiprasert, P. Efficacies of Anaerobic Microbial Consortium for Starchy Lignocellulose Hydrolysis and Acidogenic Fermentation of Cassava Pulp. Bioenerg. Res. 16, 2314–2330 (2023). https://doi.org/10.1007/s12155-023-10590-x
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-023-10590-x