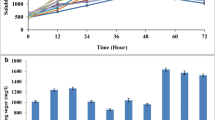
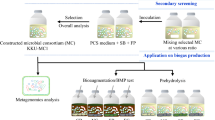
Abstract
The development of new lignocellulosic enzyme cocktails to digest raw material for the successful production of biofuels still represents great economic and scientific challenges. The aim of this study was the characterization of an aerobic consortium derived from the gut of the cockroach Nauphoeta cinerea (NaLC) under morphological, chemical, biochemical, and genetic views. Initially, the assembly of the consortium started by dissection and incubation of hindgut of N cinerea with medium containing sugarcane bagasse (BED) as carbon source. After 1 week of cultivation, the NaLC degraded about 55% of lignocellulosic material (mainly cellulose) and also promoted a significant decreased in the BED length. The biochemical analysis of consortium supernatants showed activities against Avicel, β-glucan, xylan, and CMC. The metagenomics analysis of 16S rDNA from NaLC showed 22 bacterial genomes with a predominance of Bacteroidetes (50.6%) and Proteobacteria (47.7%) with Flavobacterium spp. and Sphingomonas spp., respectively. The shotgun metagenomic analysis revealed the presence of several CAZymes, such as cellulases, hemicellulases, carbohydrate esterases, and enzymes with auxiliary activities. During the shotgun metagenomic analysis, various CAZymes organized as polysaccharide utilization loci (PUL) from Niabela sp. bacteria in NaLC were identified. It was also observed a new locus named here as LUL (lignin utilization locus) containing many genes as oxidases, dehydrogenases, peroxidases, and others that could be involved in lignin metabolization. So, this study described under different approaches an unprecedented aerobic consortium could be a useful resource of novel genes and enzymes involved in degradation of sugarcane bagasse.




Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in its Additional files.
Abbreviations
- AAs:
-
Auxiliary enzymes
- Avicel:
-
Microcrystalline cellulose
- Avicelase:
-
Enzyme that break avicel
- BED:
-
Pre-treated sugarcane bagasse
- BHB:
-
Bushnell Haas Broth commercial medium
- CAZymes:
-
Carbohydrate-active enzymes
- CBMs:
-
Carbohydrate binding modules
- CEs:
-
Carbohydrates esterases
- CMC:
-
Carboxymethylcellulose
- CMCase:
-
Enzyme that break carboxymethylcellulose
- CNPEM:
-
Centro Nacional de Pesquisa em Energia e Materiais
- DNS:
-
3,5-Dinitrosalicylic acid
- DUFs:
-
Domains of unknown function
- GHs:
-
Glycoside hydrolases
- GTs:
-
Glycosyl transferases
- HMF:
-
Furfural/hydroxymethyl furfural
- H2O2 :
-
Hydrogen peroxide
- HPAEC-PAD:
-
High-performance anion exchange with pulsed amperometric detection
- ITS:
-
Internal Transcribed Spacer 2
- LNBR:
-
Brazilian Biorenewables National Laboratory
- LULs:
-
Lignin utilization loci
- MAGs:
-
Metagenome-assembled genomes
- MnSOD:
-
Manganese superoxide dismutase
- NaLC:
-
Consortium derived from the gut of Nauphoeta cinerea
- ORFs:
-
Open reading frames
- OTU:
-
Operational taxonomic units
- PUL:
-
Polysaccharide utilization loci
References
SH Mood AH Golfeshan M Tabatabaei GS Jouzani GH Najafi M Gholami M Ardjmand 2013 Lignocellulosic biomass to bioethanol, a comprehensive review with a focus on pretreatment Renew Sustain Energy Rev 27 77 93 https://doi.org/10.1016/j.rser.2013.06.033
S Wongwilaiwalin U Rattanachomsri T Laothanachareon L Eurwilaichitr Y Igarashi V Champreda 2010 Analysis of a thermophilic lignocellulose degrading microbial consortium and multi-species lignocellulolytic enzyme system Enzyme Microb Technol 47 283 290 https://doi.org/10.1016/j.enzmictec.2010.07.013
Sun J, Zhou XJ (2011) Utilization of lignocellulose- feeding insects for viable biofuels: an emerging and promising area of entomological science. In: Liu T., Kang L. (eds) Recent Advances in Entomological Research. Springer, Berlin, Heidelberg. https://doi.org/10.1007/978-3-642-17815-3_25
C Schauer CL Thompson A Brune 2012 The bacterial community in the gut of the cockroach Shelfordella lateralis reflects the close evolutionary relatedness of cockroaches and termites Appl Environ Microbiol 78 2758 2767 https://doi.org/10.1128/AEM.07788-11
DRA Wharton ML Wharton 1965 The cellulase content of various species of cockroach J Insect Physiol 11 1401 1405 https://doi.org/10.1016/0022-1910(65)90177-0
EN Elpidina KS Vinokurov VA Gromenko YA Rudenskaya YE Dunaevsky DP Zhuzhikov 2001 Compartmentalization of proteinases and amylases in Nauphoeta cinerea midgut Arch Insect Biochem Physiol 48 206 216 https://doi.org/10.1002/arch.10000
LL Jiang JJ Zhou CS Quan ZL **u 2017 Advances in industrial microbiome based on microbial consortium for biorefinery Bioresour Bioprocess 4 1 11 https://doi.org/10.1186/s40643-017-0141-0
C Xu H Yu 2021 Insights into constructing a stable and efficient microbial consortium Chin J Chem Eng 30 112 120 https://doi.org/10.1016/j.cjche.2020.12.012
LD Bushnell HF Haas 1941 The utilization of certain hydrocarbons by microorganisms J Bacteriol 41 653 673
ER Gouveia RTD Nascimento AM Souto-Maior GJM Rocha De 2009 Validação de metodologia para a caracterização química de bagaço de cana-de-açúcar Quim Nova 32 1500 1503 https://doi.org/10.1590/S0100-40422009000600026
GJM Rocha AR Gonçalves BR Oliveira EG Olivares CEV Rossell 2012 Steam explosion pretreatment reproduction and alkaline delignification reactions performed on a pilot scale with sugarcane bagasse for bioethanol production Ind Crop Prod 35 274 279 https://doi.org/10.1016/j.indcrop.2011.07.010
A Vera-Ponce de León BC Jahnes J Duan LA Camuy-Vélez ZL Sabree 2020 Cultivable, host-specific Bacteroidetes symbionts exhibit diverse polysaccharolytic strategies Appl Environ Microbiol 86 8 1 24 https://doi.org/10.1128/AEM.00091-20
G Miller R Blum W Glennon A Burton 1960 Measurement of carboxymethylcellulase activity Anal Biochem 132 127 132 https://doi.org/10.1016/0003-2697(60)90004-X
Rasband WS (1997) Image J. Bethesda, MD National Institutes of Health. http://imagej.nih.gov/ij.
JG Caporaso CL Lauber WA Walters D Berg-Lyons J Huntley N Fierer SM Owens J Betley L Fraser M Bauer N Gormley JA Gilbert G Smith R Knight 2012 Ultra-high-throughput microbial community analysis on the Illumina HiSeq and MiSeq platforms ISME J 6 1621 1624 https://doi.org/10.1038/ismej.2012.8
White TJ, Bruns S, Lee S, Taylor J (1990) Amplification and direct sequencing of fungal ribosomal RNA genes for phylogenetics. PCR Protocols: a guide to methods and applications. In: Innis MA, Gelfand DH editors. London: Academic Press 315–322.
RC Edgar 2013 UPARSE: highly accurate OTU sequences from microbial amplicon reads Nat Methods 10 996 998 https://doi.org/10.1038/nmeth.2604
Q Wang GM Garrity JM Tiedje JR Cole 2007 Naïve Bayesian classifier for rapid assignment of rRNA sequences into the new bacterial taxonomy Appl Environ Microbiol 73 5261 5267 https://doi.org/10.1128/AEM.00062-07
RH Nilsson L Tedersoo M Ryberg E Kristiansson M Hartmann M Unterseher 2015 A comprehensive, automatically updated fungal ITS sequence dataset for reference-based chimera control in environmental sequencing efforts Microbes Environ 30 145 150 https://doi.org/10.1264/jsme2.ME14121
F Peng P Peng F Xu RC Sun 2012 Fractional purification and bioconversion of hemicelluloses Biotechnol Adv 30 879 903 https://doi.org/10.1016/j.biotechadv.2012.01.018
T Seemann 2014 Prokka: rapid prokaryotic genome annotation Bioinformatics 30 2068 2069 https://doi.org/10.1093/bioinformatics/btu153
J Alneberg BS Bjarnason I Bruijn M Schirmer J Quick UZ Ijaz L Lahti NJ Loman AF Anderson C Quince 2014 Binning metagenomic contigs by coverage and composition Nat Methods 11 1144 1146 https://doi.org/10.1038/nmeth.3103
DH Parks M Imelfort CT Skennerton P Hugenholtz GW Tyson 2015 CheckM: assessing the quality of microbial genomes recovered from isolates, single cells, and metagenomes Genome Res 25 1043 1055 https://doi.org/10.1101/gr.186072.114
PA Chaumeil AJ Mussig P Hugenholtz DH Parks 2019 GTDB-Tk: a toolkit to classify genomes with the Genome Taxonomy Database Bioinformatics 36 1925 1927 https://doi.org/10.1093/bioinformatics/btz848
Y Yin X Mao J Yang X Chen F Mao Y Xu 2012 dbCAN: a web resource for automated carbohydrate-active enzyme annotation Nucleic Acids Res 40 445 451 https://doi.org/10.1093/nar/gks479
V Lombard GH Ramulo E Drula PM Coutinho B Henrissat 2014 The carbohydrate-active enzymes database (CAZy) in 2013 Nucleic Acids Res 42 490 495 https://doi.org/10.1093/nar/gkt1178
S El-Gebali J Mistry A Bateman SR Eddy A Luciani SC Potter M Qureshi LJ Richardson GA Salazar A Smart ELL Sonnhammer L Hirsh L Paladin D Piovesan SCE Tosatto RD Finn 2019 The Pfam protein families’ database in 2019 Nucleic Acids Res 8 427 432 https://doi.org/10.1093/nar/gky995
H Zhang T Yohe L Huang S Entwistle P Wu Z Yang PK Busk Y Xu Y Yin 2018 dbCAN2: a meta server for automated carbohydrate-active enzyme annotation Nucleic Acids Res 46 95 101 https://doi.org/10.1093/nar/gky418
ME Himmel Q Xu Y Luo SY Ding R Lamed EA Bayer 2010 Microbial enzyme systems for biomass conversion: emerging paradigms Biofuels 1 323 341 https://doi.org/10.4155/bfs.09.25
SD Shinde X Meng R Kumar AJ Ragauskas 2018 Recent advances in understanding the pseudolignin formation in a lignocellulosic biorefinery Green Chem 20 2192 2205 https://doi.org/10.1039/C8GC00353J
F Hu S Jung AJ Ragauskas 2012 Pseudo-lignin formation and its impact on enzymatic hydrolysis Bioresour Technol 117 7 12 https://doi.org/10.1016/j.biortech.2012.04.037
P Kanokratana W Mhuantong T Laothanachareon S Tangphatsornruang L Eurwilaichitr K Pootanakit V Champreda 2013 Phylogenetic analysis and metabolic potential of microbial communities in an industrial bagasse collection site Microb Ecol 66 322 334 https://doi.org/10.1007/s00248-013-0209-0
D Lednická J Mergaert MC Cnockaert J Swings 2011 Isolation and identification of cellulolytic bacteria involved in the degradation of natural cellulosic fibres Syst Appl Microbiol 23 292 299 https://doi.org/10.1016/S0723-2020(00)80017-X
K Kersters De vos P, Gillis M, Swings J, Vandamme P, Stackebrandt E, 2006 Introduction to the Proteobacteria Prokaryotes 5 3 37 https://doi.org/10.1007/0-387-30745-1_1
L Auer A Lazuka D Sillam-Dusses E Miambi M O’Donohue G Hernandez Raquet 2017 Uncovering the potential of termite gut microbiome for lignocellulose bioconversion in anaerobic batch bioreactors Front Microbiol 8 2623 https://doi.org/10.3389/fmicb.2017.02623
S Jang Y Kikuchi 2020 Impact of the insect gut microbiota on ecology, evolution, and industry Curr Opin Insect Sci 41 33 39 https://doi.org/10.1016/j.cois.2020.06.004
E Mnich R Vanholme P Oyarce S Liu F Lu G Goeminne B Jørgensen 2017 Degradation of lignin b-aryl ether units in Arabidopsis thaliana expressing LigD, LigF and LigG from Sphingomonas paucimobilis SYK-6 Plant Biotechnol J 15 581 593 https://doi.org/10.1111/pbi.12655
KC Shin DK Oh 2014 Characterization of a novel recombinant β-glucosidase from Sphingopyxis alaskensis that specifically hydrolyzes the outer glucose at the C-3 position in protopanaxadiol-type ginsenosides J Biotechnol 172 30 37
NL Ward JF Challacombe PH Janssen B Henrissat PMM Coutinho G Wu DH **e M Haft J Sait RD Badger B Barabote TS Bradley LM Brettin D Brinkac T Bruce SC Creasy TM Daugherty 2009 Three genomes from the phylum Acidobacteria provide insight into the lifestyles of these microorganisms in soils Appl Environ Microbiol 75 2046 2056 https://doi.org/10.1128/AEM.02294-08
V Lombard T Bernard C Rancurel H Brumer PM Coutinho B Henrissat 2010 A hierarchical classification of polysaccharide lyases for glycogenomics Biochem J 432 3 437 444 https://doi.org/10.1042/BJ20101185
G Davies B Henrissat 1995 Structures and mechanisms of glycosyl hydrolases Structure 3 853 859 https://doi.org/10.1016/S0969-2126(01)00220-9
JLA Bras A Cartmell AL Carvalho G Vérze EA Bayer Y Vazana MA Correia JA Prates S Ratnaparkhe AB Boraston MJ Romão CM Fontes HJ Gilbert 2011 Structural insights into a unique cellulase fold and mechanism of cellulose hydrolysis PNAS 108 5237 5242 https://doi.org/10.1073/pnas.1015006108
L Wang G Zhang H Xu H **n Y Zhang 2019 Metagenomic analyses of microbial and carbohydrate-active enzymes in the rumen of holstein cows fed different forage-to-concentrate ratios Front Microbiol 10 649 https://doi.org/10.3389/fmicb.2019.00649
LL Li SR McCorkle S Monchy S Taghavi D Lelie van der 2009 Bioprospecting metagenomes: glycosyl hydrolases for converting biomass Biotechnol Biofuels 2 10 https://doi.org/10.1186/1754-6834-2-10
H Aspeborg PM Coutinho Y Wang H Brumer B Henrissat 2012 Evolution, substrate specificity and subfamily classification of glycoside hydrolase family 5 (GH5) BMC Evol Biol 12 186 https://doi.org/10.1186/1471-2148-12-186
K Mewis N Lenfant V Lombard B Henrissat 2016 Dividing the large glycoside hydrolase family 43 into subfamilies: a motivation for detailed enzyme characterization Appl Environ Microbiol 82 1686 1692 https://doi.org/10.1128/AEM.03453-15
F Warnecke P Luginbühl N Ivanovna M Ghassemian TH Richardson JT Stege 2007 Metagenomic and functional analysis of hindgut microbiota of a wood-feeding higher termite Nature 450 560 565 https://doi.org/10.1038/nature06269
BL Cantarel PM Coutinho C Rancurel T Bernard V Lombard B Henrissay 2009 The carbohydrate-active enzymes database (CAZy): an expert resource for glycogenomics Nucleic Acids Res 37 233 238 https://doi.org/10.1093/nar/gkn663
N Liu H Li MG Chevrette L Zhang L Cao H Zhou X Zhou Z Zhou PB Pope CR Currie Y Huang Q Wang 2019 Functional metagenomics reveals abundant polysaccharide degrading gene clusters and cellobiose utilization pathways within gut microbiota of a wood-feeding higher termite ISME J 13 104 117 https://doi.org/10.1038/s41396-018-0255-1
A Levasseur E Drula V Lombard PM Coutinho B Henrissat 2013 Expansion of the enzymatic repertoire of the CAZy database to integrate auxiliary redox enzymes Biotechnol Biofuels 6 41 https://doi.org/10.1186/1754-6834-6-41
JP Silva ARP Ticona PRV Hamann BF Quirino EF Noronha 2021 Deconstruction of lignin: from enzymes to microorganisms Molecules 26 8 2299 https://doi.org/10.3390/molecules26082299
N Terrapon V Lombard HJ Gilbert B Henrissat 2015 Automatic prediction of polysaccharide utilization loci in Bacteroidetes species Bioinformatics 31 5 647 655 https://doi.org/10.1093/bioinformatics/btu716
JM Grondin K Tamura G Déjean DW Abbott H Brumer 2017 Polysaccharide utilization loci: fueling microbial communities J Bacteriol 199 e00860 e916 https://doi.org/10.1128/JB.00860-16
MH Foley DW Cockburn NM Koropatkin 2016 The Sus operon: a model system for starch uptake by the human gut Bacteroidetes Cell Mol Life Sci 73 2603 2617 https://doi.org/10.1007/s00018-016-2242-x
L Sützl CVFP Laurent AT Abrera G Schütz R Ludwig D Haltrich 2018 Multiplicity of enzymatic functions in the CAZy AA3 family Appl Microbiol Biotechnol 102 2477 2492 https://doi.org/10.1007/s00253-018-8784-0
L Lin Y Cheng Y Pu S Sun X Li M ** EA Pierson DC Gross BE Dale SY Dai AJ Ragauskas JS Yuan 2016 Systems biology-guided biodesign of consolidated lignin conversion Green Chem 18 20 5536 5547 https://doi.org/10.1039/C6GC01131D
GMM Rashid CR Taylor Y Liu X Zhang D Rea V Fülöp TDH Bugg 2015 Identification of manganese superoxide dismutase from Sphingobacterium sp. T2 as a novel bacterial enzyme for lignin oxidation ACS Chem Biol 10 2286 2294 https://doi.org/10.1021/acschembio.5b00298
Acknowledgements
We would like to express our gratitude to David Forrest for editorial assistance.
Funding
This work was supported by grants from the Coordenação de Aperfeiçoamento de Pessoal de Nível Superior (CAPES), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro (FAPERJ), Fundação de Amparo à Pesquisa do Estado de São Paulo (Fapesp). Conselho Nacional de Desenvolvimento Científico e Tecnológico (CNPq), and INCT-Entomologia Molecular.
Author information
Authors and Affiliations
Contributions
D.B.C. maintained bacteria consortia in laboratory conditions, performed experiments, analyzed data, edited figures, created the line drawing, and drafted the manuscript; D.A.P. maintained bacteria consortia in laboratory conditions, performed experiments, and analyzed data; G.F.P. performed the bioinformatics analysis of the article; J.C. performed and supervised the experiments of enzymatic activities; S.C.R. performed the chemical analysis of the article; A.G. performed the chemical and statistical analysis of the article; A.P.S. performed the chemical and statistical analysis of the article; M.B. performed the chemical and statistical analysis of the article; R.R. performed the sequencing experiments and its analysis; L.M.Z. performed the sequencing experiments and its analysis; F.M.S. designed, provided financial support, analyzed data; J.P.L.C. conceived, designed, and coordinated the study, analyzed data, and reviewed the manuscript; E.A.M conceived, designed, and coordinated the study, provided financial support, analyzed data, and reviewed the manuscript. All authors gave final approval for publication.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Carvalho, D.B., Paixão, D.A., Persinoti, G.F. et al. Degradation of Sugarcane Bagasse by Cockroach Consortium Bacteria. Bioenerg. Res. 15, 1144–1156 (2022). https://doi.org/10.1007/s12155-021-10363-4
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12155-021-10363-4