Abstract
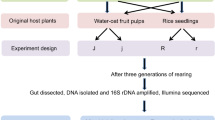
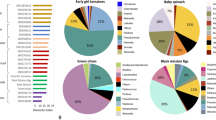
Oxycarenus laetus is a cotton pest that primarily feeds on seeds that are rich in gossypol. Though gossypol is toxic to general herbivores, O. laetus does not show such complications and instead grows and reproduces well on cotton plants compared to its other hosts. In this study, we have fed O. laetus with natural and induced gossypol-based diets to explore the difference in its gut microbiota. We performed NGS 16S rRNA amplicon sequencing on the Illumina MiSeq platform and analyzed the data using the QIIME2 pipeline supplemented with Greengenes and EZBioCloud reference databases. We also used culture-based methods to identify a few abundant gut bacteria present in O. laetus. Enterococcus faecalis, Wolbachia bourtzisii, Wolbachia pipientis, Corynebacterium glyciniphilum, Staphylococcus sciuri, and Kocuria rosea were some of the major species that formed the core gut microbiome of O. laetus. We have also observed that some species were present only in the sample with the highest concentration of gossypol, signifying that they might have the potential to degrade gossypol.

Similar content being viewed by others
References
Wink M (2018) Plant secondary metabolites modulate insect behavior-steps toward addiction? Front Physiol 9:1–9. https://doi.org/10.3389/fphys.2018.00364
Cui S, Wang L, Ma L, Geng X (2016) P450-mediated detoxification of botanicals in insects. Phytoparasitica 44:585–599. https://doi.org/10.1007/s12600-016-0550-1
Wu C, Chakrabarty S, ** M et al (2019) Insect ATP-binding cassette (ABC) transporters: Roles in xenobiotic detoxification and Bt insecticidal activity. Int J Mol Sci 20:12–14. https://doi.org/10.3390/ijms20112829
Berasategui A, Salem H, Paetz C et al (2017) Gut microbiota of the pine weevil degrades conifer diterpenes and increases insect fitness. Mol Ecol 26:4099–4110. https://doi.org/10.1111/mec.14186
Zhang S, Shu J, Xue H et al (2020) The gut microbiota in camellia weevils are influenced by plant secondary metabolites and contribute to saponin degradation. mSystems 5:1–17. https://doi.org/10.1128/msystems.00692-19
Iqbal J, Bhutta SA, Alqarni AS et al (2018) Seasonal population dynamics of dusky cotton bug (Oxycarenus spp.) in transgenic cotton varieties under field conditions. Saudi J Biol Sci 25:1122–1127. https://doi.org/10.1016/j.sjbs.2017.05.004
Krempl C, Sporer T, Reichelt M et al (2016) Potential detoxification of gossypol by UDP-glycosyltransferases in the two Heliothine moth species Helicoverpa armigera and Heliothis virescens. Insect Biochem Mol Biol 71:49–57. https://doi.org/10.1016/j.ibmb.2016.02.005
Andrews S (2010) FastQC: a quality control tool for high throughput sequence data. http://www.bioinformatics.babraham.ac.uk/projects/fastqc/. Accessed 21 Dec 2018
Bolyen E, Rideout JR, Dillon MR et al (2019) Reproducible, interactive, scalable and extensible microbiome data science using QIIME 2. Nat Biotechnol 37:852–857. https://doi.org/10.1038/s41587-019-0209-9
Martin M (2011) Cutadapt removes adapter sequences from high-throughput sequencing reads. EMBnet J 17(1):10–12. https://doi.org/10.14806/ej.17.1.200
Callahan BJ, McMurdie PJ, Rosen MJ et al (2016) DADA2: High-resolution sample inference from Illumina amplicon data. Nat Methods 13:581–583. https://doi.org/10.1038/nmeth.3869
Rognes T, Flouri T, Nichols B et al (2016) VSEARCH: A versatile open source tool for metagenomics. PeerJ 2016:1–22. https://doi.org/10.7717/peerj.2584
DeSantis TZ, Hugenholtz P, Larsen N et al (2006) Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl Environ Microbiol 72:5069–5072. https://doi.org/10.1128/AEM.03006-05
Yoon SH, Ha SM, Kwon S et al (2017) Introducing EzBioCloud: a taxonomically united database of 16S rRNA gene sequences and whole-genome assemblies. Int J Syst Evol Microbiol 67:1613–1617. https://doi.org/10.1099/ijsem.0.001755
Kumar S, Stecher G, Li M et al (2018) MEGA X: molecular evolutionary genetics analysis across computing platforms. Mol Biol Evol 35:1547–1549. https://doi.org/10.1093/molbev/msy096
Priya NG, Ojha A, Kajla MK et al (2012) Host plant induced variation in gut bacteria of Helicoverpa armigera. PLoS ONE 7:1–10. https://doi.org/10.1371/journal.pone.0030768
Klammsteiner T, Walter A, Bogataj T et al (2020) The core gut microbiome of black soldier fly (Hermetia illucens) larvae raised on low-bioburden diets. Front Microbiol 11:1–14. https://doi.org/10.3389/fmicb.2020.00993
Shinde AA, Shaikh FK, Gadge PP et al (2019) Conserved nature of Helicoverpa armigera gut bacterial flora on different host plants and in vitro interactions with PI proteins advocates role in host digestive physiology. J Saudi Soc Agric Sci 18:141–149. https://doi.org/10.1016/j.jssas.2017.03.004
Schmid RB, Lehman RM, Brözel VS, Lundgren JG (2014) An indigenous gut bacterium, Enterococcus faecalis (Lactobacillales: Enterococcaceae), increases seed consumption by Harpalus pensylvanicus (Coleoptera : Carabidae). Florida Entomol 97:575–584. https://doi.org/10.1653/024.097.0232
Saurav GK, Daimei G, Rana VS et al (2016) Detection and localization of Wolbachia in Thrips palmi Karny (Thysanoptera: Thripidae). Indian J Microbiol 56:167–171. https://doi.org/10.1007/s12088-016-0567-7
Hosokawa T, Koga R, Kikuchi Y et al (2010) Wolbachia as a bacteriocyte-associated nutritional mutualist. Proc Natl Acad Sci USA 107:769–774. https://doi.org/10.1073/pnas.0911476107
Risa A, Krifaton C, Kukolya J et al (2018) Aflatoxin B1 and zearalenone-detoxifying profile of Rhodococcus type strains. Curr Microbiol 75:907–917. https://doi.org/10.1007/s00284-018-1465-5
Pan L, Gu JG, Yin B, Cheng SP (2009) Contribution to deterioration of polymeric materials by a slow-growing bacterium Nocardia corynebacterioides. Int Biodeterior Biodegrad 63:24–29. https://doi.org/10.1016/j.ibiod.2008.06.003
Bayat Z, Hassanshahian M, Cappello S (2015) Immobilization of microbes for bioremediation of crude oil polluted environments: A mini review. Open Microbiol J 9:48–54. https://doi.org/10.2174/1874285801509010048
Ozdal M, Ozdal OG, Algur OF (2016) Isolation and characterization of α-endosulfan degrading bacteria from the microflora of cockroaches. Polish J Microbiol 65:63–68. https://doi.org/10.5604/17331331.1197325
Acknowledgements
A part of this study is funded by DST – SERB, India under the young scientist scheme: file#- YSS/2014/000293. Authors thank the administration and management of SRM Institute of Science and Technology for all the support provided.
Author information
Authors and Affiliations
Corresponding author
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Sureshan, S.C., Mohideen, H.S. & Nair, T.S. Gut Metagenomic Profiling of Gossypol Induced Oxycarenus laetus (Hemiptera: Lygaeidae) Reveals Gossypol Tolerating Bacterial Species. Indian J Microbiol 62, 54–60 (2022). https://doi.org/10.1007/s12088-021-00964-0
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12088-021-00964-0