Abstract
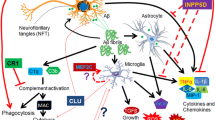
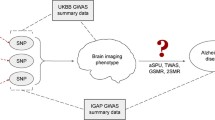
Previous in vitro and post-mortem studies have reported the role of inflammation in neurodegenerative disorders. However, the association between inflammation and brain structure in vivo and the transcriptome-driven functional basis with relevance to neurodegenerative disorders remains elusive. The aim of the present study is to identify the association among inflammation, brain structure, and neurodegenerative disorders at genetic and transcriptomic levels. Genetic variants associated with inflammatory cytokines were selected from the latest and largest genome-wide association studies of European ancestry. Neurodegenerative disorders including Alzheimer’s disease (AD), Parkinson’s disease (PD), amyotrophic lateral sclerosis (ALS), and dementia with Lewy bodies (DLB) and brain structure imaging measures were selected as the outcomes. Two-sample Mendelian randomization analyses were conducted to identify the causal associations. Single-nucleus transcriptome data of the occipitotemporal cortex was further analyzed to identify the differential expressed genes in AD, which were tested for biological processes and protein interaction network. MR analysis indicated that genetically predicted TREM2 and sTREM2 were significantly associated with AD (TREM2: z-score = −9.088, p-value = 1.02 × 10−19; sTREM2: z-score = −7.495, p-value = 6.61 × 10−14). The present study found no evidence to support the causal associations between other inflammatory cytokines and the risks of AD, PD, ALS, or DLB. Genetically predicted TREM2 was significantly associated with the cortical thickness of inferior temporal (z-score = −4.238, p-value = 2.26 × 10−5) and pole temporal (z-score = −4.549, p-value = 5.40 × 10−6). In the occipitotemporal cortex samples, microglia were the main source of TREM2 gene and showed increasing expression of genes associated with inflammation and immunity. The present study has leveraged genetic and transcriptomic data to identify the association among TREM2, temporal lobe, and AD and the underlying cellular and molecular basis, thus providing a new perspective on the role of TREM2 in AD and insights into the complex associations among inflammation, brain structure, and neurodegenerative disorders, particularly AD.





Similar content being viewed by others
Data Availability
All data used in the present study were publicly available. The summary-level GWAS datasets of inflammatory cytokines and neurodegenerative disorders were acquired from GWAS Catalog website (https://www.ebi.ac.uk/gwas). The summary-level GWAS datasets of brain structure imaging measures were acquired from Oxford Brain Imaging Genetics Server BIG40 (https://open.win.ox.ac.uk/ukbiobank/big40/).
Abbreviations
- AD:
-
Alzheimer’s disease
- ALS:
-
Amyotrophic lateral sclerosis
- CRP:
-
C-reactive protein
- CSF:
-
Cerebrospinal fluid
- CTR:
-
Control
- CX3CL1:
-
Fractalkine
- DEG:
-
Differentially expressed gene
- DLB:
-
Dementia due to Lewy’s bodies
- GO:
-
Gene Ontology
- GSEA:
-
Gene-set enrichment analysis
- GWAS:
-
Genome-wide association study
- IL-1:
-
Interleukin-1
- IL-2:
-
Interleukin-2
- IL-6:
-
Interleukin-6
- IVW:
-
Inverse-variance weighted
- KEGG:
-
Kyoto Encyclopedia of Genes and Genomes
- MR:
-
Mendelian randomization
- MSigDB:
-
Molecular Signatures Database
- PD:
-
Parkinson’s disease
- TREM2:
-
Triggering receptor expressed on myeloid cell 2
- TNFR1:
-
Tumor necrosis factor receptor 1
- TNFR2:
-
Tumor necrosis factor receptor 2
- TNFSF14:
-
Tumor necrosis factor ligand superfamily member 14
- TRAIL:
-
TNF-related apoptosis-inducing ligand
- TRAILR2:
-
TNF-related apoptosis-inducing ligand receptor 2
- TRANCE:
-
TNF-related activation-induced cytokine
- UKB:
-
UK Biobank
- YKL40:
-
Chitinase-3-like protein 1
References
Glass CK, Saijo K, Winner B et al (2010) Mechanisms underlying inflammation in neurodegeneration. Cell 140:918–934. https://doi.org/10.1016/j.cell.2010.02.016
Heneka MT, Carson MJ, El Khoury J et al (2015) Neuroinflammation in Alzheimer’s disease. Lancet Neurol 14:388–405. https://doi.org/10.1016/S1474-4422(15)70016-5
Hirsch EC, Hunot S (2009) Neuroinflammation in Parkinson’s disease: a target for neuroprotection? Lancet Neurol 8:382–397. https://doi.org/10.1016/S1474-4422(09)70062-6
McCauley ME, Baloh RH (2019) Inflammation in ALS/FTD pathogenesis. Acta Neuropathol 137:715–730. https://doi.org/10.1007/s00401-018-1933-9
Amin J, Erskine D, Donaghy PC et al (2022) Inflammation in dementia with Lewy bodies. Neurobiol Dis 168:105698. https://doi.org/10.1016/j.nbd.2022.105698
Leng F, Edison P (2021) Neuroinflammation and microglial activation in Alzheimer disease: where do we go from here? Nat Rev Neurol 17:157–172. https://doi.org/10.1038/s41582-020-00435-y
Shi Y, Holtzman DM (2018) Interplay between innate immunity and Alzheimer disease: APOE and TREM2 in the spotlight. Nat Rev Immunol 18:759–772. https://doi.org/10.1038/s41577-018-0051-1
Cheng X, Shen Y, Li R (2014) Targeting TNF: a therapeutic strategy for Alzheimer’s disease. Drug Discov Today 19:1822–1827. https://doi.org/10.1016/j.drudis.2014.06.029
Luan Y-Y, Yao Y-M (2018) The clinical significance and potential role of C-reactive protein in chronic inflammatory and neurodegenerative diseases. Front Immunol 9:1302. https://doi.org/10.3389/fimmu.2018.01302
Green C, Shen X, Stevenson AJ et al (2021) Structural brain correlates of serum and epigenetic markers of inflammation in major depressive disorder. Brain Behav Immun 92:39–48. https://doi.org/10.1016/j.bbi.2020.11.024
Williams JA, Burgess S, Suckling J et al (2022) Inflammation and brain structure in schizophrenia and other neuropsychiatric disorders: a Mendelian randomization study. JAMA Psychiatry 79:498–507. https://doi.org/10.1001/jamapsychiatry.2022.0407
MacKenzie G, Subramaniam S, Caldwell LJ et al (2021) Research priorities for neuroimmunology: identifying the key research questions to be addressed by 2030. Wellcome Open Res 6:194. https://doi.org/10.12688/wellcomeopenres.16997.1
Ransohoff RM (2016) How neuroinflammation contributes to neurodegeneration. Science 353:777–783. https://doi.org/10.1126/science.aag2590
Lawlor DA, Harbord RM, Sterne JAC et al (2008) Mendelian randomization: using genes as instruments for making causal inferences in epidemiology. Stat Med 27:1133–1163. https://doi.org/10.1002/sim.3034
Davies NM, Holmes MV, Davey Smith G (2018) Reading Mendelian randomisation studies: a guide, glossary, and checklist for clinicians. BMJ 362:k601. https://doi.org/10.1136/bmj.k601
Wang M, Song W-M, Ming C et al (2022) Guidelines for bioinformatics of single-cell sequencing data analysis in Alzheimer’s disease: review, recommendation, implementation and application. Mol Neurodegener 17:17. https://doi.org/10.1186/s13024-022-00517-z
Gerrits E, Brouwer N, Kooistra SM et al (2021) Distinct amyloid-β and tau-associated microglia profiles in Alzheimer’s disease. Acta Neuropathol 141:681–696. https://doi.org/10.1007/s00401-021-02263-w
Finneran DJ, Nash KR (2019) Neuroinflammation and fractalkine signaling in Alzheimer’s disease. J Neuroinflammation 16:30. https://doi.org/10.1186/s12974-019-1412-9
Nordengen K, Kirsebom B-E, Henjum K et al (2019) Glial activation and inflammation along the Alzheimer’s disease continuum. J Neuroinflammation 16:46. https://doi.org/10.1186/s12974-019-1399-2
Herder C, Nuotio M-L, Shah S et al (2014) Genetic determinants of circulating interleukin-1 receptor antagonist levels and their association with glycemic traits. Diabetes 63:4343–4359. https://doi.org/10.2337/db14-0731
Ahola-Olli AV, Würtz P, Havulinna AS et al (2017) Genome-wide association study identifies 27 loci influencing concentrations of circulating cytokines and growth factors. Am J Hum Genet 100:40–50. https://doi.org/10.1016/j.ajhg.2016.11.007
Folkersen L, Gustafsson S, Wang Q et al (2020) Genomic and drug target evaluation of 90 cardiovascular proteins in 30,931 individuals. Nat Metab 2:1135–1148. https://doi.org/10.1038/s42255-020-00287-2
Ligthart S, Vaez A, Võsa U et al (2018) Genome analyses of >200,000 individuals identify 58 loci for chronic inflammation and highlight pathways that link inflammation and complex disorders. Am J Hum Genet 103:691–706. https://doi.org/10.1016/j.ajhg.2018.09.009
Gudjonsson A, Gudmundsdottir V, Axelsson GT et al (2022) A genome-wide association study of serum proteins reveals shared loci with common diseases. Nat Commun 13. https://doi.org/10.1038/s41467-021-27850-z
Hou X-H, Bi Y-L, Tan M-S et al (2019) Genome-wide association study identifies Alzheimer’s risk variant in MS4A6A influencing cerebrospinal fluid sTREM2 levels. Neurobiol Aging 84:241.e13–241.e20. https://doi.org/10.1016/j.neurobiolaging.2019.05.008
Schwartzentruber J, Cooper S, Liu JZ et al (2021) Genome-wide meta-analysis, fine-map**, and integrative prioritization implicate new Alzheimer’s disease risk genes. Nat Genet 53:392–402. https://doi.org/10.1038/s41588-020-00776-w
Nalls MA, Blauwendraat C, Vallerga CL et al (2019) Identification of novel risk loci, causal insights, and heritable risk for Parkinson’s disease: a meta-analysis of genome-wide association studies. Lancet Neurol 18:1091–1102. https://doi.org/10.1016/S1474-4422(19)30320-5
van Rheenen W, van der Spek RAA, Bakker MK et al (2021) Common and rare variant association analyses in amyotrophic lateral sclerosis identify 15 risk loci with distinct genetic architectures and neuron-specific biology. Nat Genet 53:1636–1648. https://doi.org/10.1038/s41588-021-00973-1
Chia R, Sabir MS, Bandres-Ciga S et al (2021) Genome sequencing analysis identifies new loci associated with Lewy body dementia and provides insights into its genetic architecture. Nat Genet 53:294–303. https://doi.org/10.1038/s41588-021-00785-3
Smith SM, Douaud G, Chen W et al (2021) An expanded set of genome-wide association studies of brain imaging phenotypes in UK Biobank. Nat Neurosci 24:737–745. https://doi.org/10.1038/s41593-021-00826-4
Burgess S, Butterworth A, Thompson SG (2013) Mendelian randomization analysis with multiple genetic variants using summarized data. Genet Epidemiol 37:658–665. https://doi.org/10.1002/gepi.21758
Bowden J, Del Greco MF, Minelli C et al (2017) A framework for the investigation of pleiotropy in two-sample summary data Mendelian randomization. Stat Med 36:1783–1802. https://doi.org/10.1002/sim.7221
Bowden J, Davey Smith G, Burgess S (2015) Mendelian randomization with invalid instruments: effect estimation and bias detection through Egger regression. Int J Epidemiol 44:512–525. https://doi.org/10.1093/ije/dyv080
Hemani G, Zheng J, Elsworth B et al (2018) The MR-Base platform supports systematic causal inference across the human phenome. Elife 7:e34408. https://doi.org/10.7554/eLife.34408
Butler A, Hoffman P, Smibert P et al (2018) Integrating single-cell transcriptomic data across different conditions, technologies, and species. Nat Biotechnol 36:411–420. https://doi.org/10.1038/nbt.4096
Korsunsky I, Millard N, Fan J et al (2019) Fast, sensitive and accurate integration of single-cell data with Harmony. Nat Methods 16:1289–1296. https://doi.org/10.1038/s41592-019-0619-0
Yu G, Wang L-G, Han Y, He Q-Y (2012) clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 16:284–287. https://doi.org/10.1089/omi.2011.0118
Wu T, Hu E, Xu S et al (2021) clusterProfiler 4.0: a universal enrichment tool for interpreting omics data. Innovation (Camb) 2:100141. https://doi.org/10.1016/j.xinn.2021.100141
Liberzon A, Birger C, Thorvaldsdóttir H et al (2015) The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst 1:417–425. https://doi.org/10.1016/j.cels.2015.12.004
Yang J, Fu Z, Zhang X et al (2020) TREM2 ectodomain and its soluble form in Alzheimer’s disease. J Neuroinflammation 17(1):204. https://doi.org/10.1186/s12974-020-01878-2
Yeh FL, Hansen DV, Sheng M (2017) TREM2, microglia, and neurodegenerative diseases. Trends Mol Med 23:512–533. https://doi.org/10.1016/j.molmed.2017.03.008
Takahashi K, Rochford CDP, Neumann H (2005) Clearance of apoptotic neurons without inflammation by microglial triggering receptor expressed on myeloid cells-2. J Exp Med 201:647–657. https://doi.org/10.1084/jem.20041611
Zhu B, Liu Y, Hwang S et al (2022) Trem2 deletion enhances tau dispersion and pathology through microglia exosomes. Mol Neurodegener 17:58. https://doi.org/10.1186/s13024-022-00562-8
Filipello F, Goldsbury C, You SF et al (2022) Soluble TREM2: innocent bystander or active player in neurological diseases? Neurobiol Dis 165:105630. https://doi.org/10.1016/j.nbd.2022.105630
Wang Y, Cella M, Mallinson K et al (2015) TREM2 lipid sensing sustains the microglial response in an Alzheimer’s disease model. Cell 160:1061–1071. https://doi.org/10.1016/j.cell.2015.01.049
Zhong L, Xu Y, Zhuo R et al (2019) Soluble TREM2 ameliorates pathological phenotypes by modulating microglial functions in an Alzheimer’s disease model. Nat Commun 10:1365. https://doi.org/10.1038/s41467-019-09118-9
Tan YJ, Ng ASL, Vipin A et al (2017) Higher peripheral TREM2 mRNA levels relate to cognitive deficits and hippocampal atrophy in Alzheimer’s disease and amnestic mild cognitive impairment. J Alzheimers Dis 58:413–423. https://doi.org/10.3233/JAD-161277
Falcon C, Monté-Rubio GC, Grau-Rivera O et al (2019) CSF glial biomarkers YKL40 and sTREM2 are associated with longitudinal volume and diffusivity changes in cognitively unimpaired individuals. Neuroimage Clin 23:101801. https://doi.org/10.1016/j.nicl.2019.101801
Halaas NB, Henjum K, Blennow K et al (2020) CSF sTREM2 and tau work together in predicting increased temporal lobe atrophy in older adults. Cereb Cortex 30:2295–2306. https://doi.org/10.1093/cercor/bhz240
Samanci B, Bilgiç B, Gelişin Ö et al (2021) TREM2 variants as a possible cause of frontotemporal dementia with distinct neuroimaging features. Eur J Neurol 28:2603–2613. https://doi.org/10.1111/ene.14908
Suárez-Calvet M, Kleinberger G, Araque Caballero MÁ et al (2016) sTREM2 cerebrospinal fluid levels are a potential biomarker for microglia activity in early-stage Alzheimer’s disease and associate with neuronal injury markers. EMBO Mol Med 8:466–476. https://doi.org/10.15252/emmm.201506123
Jay TR, Hirsch AM, Broihier ML et al (2017) Disease Progression-dependent effects of TREM2 deficiency in a mouse model of Alzheimer’s disease. J Neurosci 37:637–647. https://doi.org/10.1523/JNEUROSCI.2110-16.2016
Keren-Shaul H, Spinrad A, Weiner A et al (2017) A unique microglia type associated with restricting development of Alzheimer’s disease. Cell 169. https://doi.org/10.1016/j.cell.2017.05.018
Lue L-F, Schmitz CT, Serrano G et al (2015) TREM2 protein expression changes correlate with Alzheimer’s disease neurodegenerative pathologies in post-mortem temporal cortices. Brain Pathol 25:469–480. https://doi.org/10.1111/bpa.12190
Yang L, Zhou R, Tong Y et al (2020) Neuroprotection by dihydrotestosterone in LPS-induced neuroinflammation. Neurobiol Dis 140. https://doi.org/10.1016/j.nbd.2020.104814
Qu L, Pan C, He S-M et al (2019) The Ras superfamily of small GTPases in non-neoplastic cerebral diseases. Front Mol Neurosci 12:121. https://doi.org/10.3389/fnmol.2019.00121
Luo Q, Schnöder L, Hao W et al (2022) p38α-MAPK-deficient myeloid cells ameliorate symptoms and pathology of APP-transgenic Alzheimer’s disease mice. Aging Cell 21:e13679. https://doi.org/10.1111/acel.13679
Hou J, Chen Y, Grajales-Reyes G, Colonna M (2022) TREM2 dependent and independent functions of microglia in Alzheimer’s disease. Mol Neurodegener 17:84. https://doi.org/10.1186/s13024-022-00588-y
Leyns CEG, Ulrich JD, Finn MB et al (2017) TREM2 deficiency attenuates neuroinflammation and protects against neurodegeneration in a mouse model of tauopathy. Proc Natl Acad Sci U S A 114:11524–11529. https://doi.org/10.1073/pnas.1710311114
Silvin A, Uderhardt S, Piot C et al (2022) Dual ontogeny of disease-associated microglia and disease inflammatory macrophages in aging and neurodegeneration. Immunity 55:1448–1465.e6. https://doi.org/10.1016/j.immuni.2022.07.004
Deming Y, Filipello F, Cignarella F et al (2019) The MS4A gene cluster is a key modulator of soluble TREM2 and Alzheimer’s disease risk. Sci Transl Med 11:eaau2291. https://doi.org/10.1126/scitranslmed.aau2291
Ma J, Yu J-T, Tan L (2015) MS4A cluster in Alzheimer’s disease. Mol Neurobiol 51:1240–1248. https://doi.org/10.1007/s12035-014-8800-z
Acknowledgements
The authors gratefully thanked all the studies or consortia mentioned in the present study for providing summary-level GWAS data and expression matrix of single-nucleus transcriptome.
Funding
This work was funded by the Science and Technology Innovation 2030 Major Projects (2022ZD0211600), the National Natural Science Foundation of China (92249305, 82071201, 82071997), the Shanghai Municipal Science and Technology Major Project (2018SHZDZX01), the Research Start-up Fund of Huashan Hospital (2022QD002), the Excellence 2025 Talent Cultivation Program at Fudan University (3030277001), the Shanghai Talent Development Funding for the Project (2019074), the ZHANGJIANG Lab, the Tianqiao and Chrissy Chen Institute, and the State Key Laboratory of Neurobiology and Frontiers Center for Brain Science of Ministry of Education, Fudan University.
Author information
Authors and Affiliations
Contributions
JTY conceptualized the study and revised the manuscript. WSL, YRZ, WC, and YJG analyzed and interpreted the data. WSL and HFW prepared all the figures and tables. WSL and YRZ drafted and revised the manuscript. All authors contributed to the writing and revisions of the paper and approved the final version.
Corresponding author
Ethics declarations
Ethics Approval
The studies included in the present study have received ethics approval from the respective institutional review boards.
Consent to Participate
Informed consent was obtained from all individual participants included in the GWAS study and single-nucleus transcriptomic study used in the present study.
Consent for Publication
All data in this study were obtained from the published GWAS studies or single-nucleus transcriptomic study, and the participants had signed informed consent in the published original studies.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher’s Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
ESM 1
(DOCX 1222 kb)
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Liu, WS., Zhang, YR., Ge, YJ. et al. Inflammation and Brain Structure in Alzheimer’s Disease and Other Neurodegenerative Disorders: a Mendelian Randomization Study. Mol Neurobiol 61, 1593–1604 (2024). https://doi.org/10.1007/s12035-023-03648-6
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12035-023-03648-6