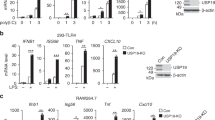
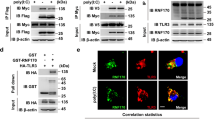
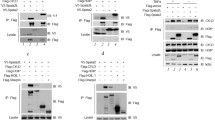
Abstract
Ubiquitination (or ubiquitylation) and de-ubiquitination, which are both post-translational modifications (PTMs) of proteins, have become a research hotspot in recent years. Some ubiquitinated or de-ubiquitinated signaling proteins have been found to promote or suppress innate immunity through Toll-like receptor (TLR), RIG-like receptor (RIG-I-like receptor, RLR), NOD-like receptor (NLR), and the cyclic guanosine monophosphate (GMP)-adenosine monophosphate (AMP) synthase (cGAS)-STING pathway. This article aimed to provide a review on the role of ubiquitination and de-ubiquitination, especially ubiquitin ligase enzymes and de-ubiquitinating enzymes, in the above four pathways. We hope that our work can contribute to the research and development of treatment strategies for innate immunity–related diseases such as inflammatory bowel disease.


Similar content being viewed by others
References
Vijay-Kumar S, Bugg CE, Cook WJ. Structure of ubiquitin refined at 1.8 A resolution. J Mol Biol. 1987;194(3):531–44. https://doi.org/10.1016/0022-2836(87)90679-6.
Hershko A, Ciechanover A. The ubiquitin system. Ann Rev Biochem. 1998;67:425–79. https://doi.org/10.1146/annurev.biochem.67.1.425.
Akutsu M, Dikic I, Bremm A. Ubiquitin chain diversity at a glance. J Cell Sci. 2016;129(5):875–80. https://doi.org/10.1242/jcs.183954.
Yau R, Rape M. The increasing complexity of the ubiquitin code. Nat Cell Biol. 2016;18(6):579–86. https://doi.org/10.1038/ncb3358.
Komander D, Rape M. The ubiquitin code. Ann Rev Biochem. 2012;81:203–29. https://doi.org/10.1146/annurev-biochem-060310-170328.
Grabbe C, Husnjak K, Dikic I. The spatial and temporal organization of ubiquitin networks. Nat Rev Mol Cell Biol. 2011;12(5):295–307. https://doi.org/10.1038/nrm3099.
Snyder NA, Silva GM. Deubiquitinating enzymes (DUBs): Regulation, homeostasis, and oxidative stress response. J Biol Chem. 2021;297(3):101077. https://doi.org/10.1016/j.jbc.2021.101077.
Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124(4):783–801. https://doi.org/10.1016/j.cell.2006.02.015.
Faenza I, Blalock WL. Innate Immunity: A Balance between Disease and Adaption to Stress. Biomolecules. 2022;12:5. https://doi.org/10.3390/biom12050737.
Tao S, Drexler I. Targeting Autophagy in Innate Immune Cells: Angel or Demon During Infection and Vaccination? Front Immunol. 2020;11:460. https://doi.org/10.3389/fimmu.2020.00460.
Takeuchi O, Akira S. Pattern recognition receptors and inflammation. Cell. 2010;140(6):805–20. https://doi.org/10.1016/j.cell.2010.01.022.
Kawai T. Akira S. SnapShot: Pattern-recognition receptors. Cell. 2007;129(5):1024. https://doi.org/10.1016/j.cell.2007.05.017.
Kumar V. Toll-like receptors in sepsis-associated cytokine storm and their endogenous negative regulators as future immunomodulatory targets. Int Immunopharmacol. 2020;89 Pt B:107087. https://doi.org/10.1016/j.intimp.2020.107087.
Brisse M, Ly H. Comparative Structure and Function Analysis of the RIG-I-Like Receptors: RIG-I and MDA5. Front Immunol. 2019;10:1586. https://doi.org/10.3389/fimmu.2019.01586.
Uhlén M, Fagerberg L, Hallström BM, Lindskog C, Oksvold P, Mardinoglu A, et al. Proteomics. Tissue-based map of the human proteome. Science. 2015;347 6220:1260419; https://doi.org/10.1126/science.1260419.
Meunier E, Broz P. Evolutionary Convergence and Divergence in NLR Function and Structure. Trends Immunol. 2017;38(10):744–57. https://doi.org/10.1016/j.it.2017.04.005.
Broz P, Dixit VM. Inflammasomes: mechanism of assembly, regulation and signalling. Nat Rev Immunol. 2016;16(7):407–20. https://doi.org/10.1038/nri.2016.58.
Ding C, Song Z, Shen A, Chen T, Zhang A. Small molecules targeting the innate immune cGAS-STING-TBK1 signaling pathway. Acta pharmaceutica Sinica B. 2020;10(12):2272–98. https://doi.org/10.1016/j.apsb.2020.03.001.
Rytkönen A, Holden DW. Bacterial interference of ubiquitination and deubiquitination. Cell Host Microbe. 2007;1(1):13–22. https://doi.org/10.1016/j.chom.2007.02.003.
Toma-Fukai S, Shimizu T. Structural Diversity of Ubiquitin E3 Ligase. Molecules (Basel, Switzerland). 2021;26:21. https://doi.org/10.3390/molecules26216682.
Rolfe M, Beer-Romero P, Glass S, Eckstein J, Berdo I, Theodoras A, et al. Reconstitution of p53-ubiquitinylation reactions from purified components: the role of human ubiquitin-conjugating enzyme UBC4 and E6-associated protein (E6AP). Proc Natl Acad Sci U S A. 1995;92(8):3264–8. https://doi.org/10.1073/pnas.92.8.3264.
Bernassola F, Chillemi G, Melino G. HECT-Type E3 Ubiquitin Ligases in Cancer. Trends Biochem Sci. 2019;44(12):1057–75. https://doi.org/10.1016/j.tibs.2019.08.004.
Lorenz S. Structural mechanisms of HECT-type ubiquitin ligases. Biol Chem. 2018;399(2):127–45. https://doi.org/10.1515/hsz-2017-0184.
Yin Q, Wyatt CJ, Han T, Smalley KSM, Wan L. ITCH as a potential therapeutic target in human cancers. Semin Cancer Biol. 2020;67(Pt 2):117–30. https://doi.org/10.1016/j.semcancer.2020.03.003.
Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, et al. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6(10):981–8. https://doi.org/10.1038/ni1243.
You F, Sun H, Zhou X, Sun W, Liang S, Zhai Z, et al. PCBP2 mediates degradation of the adaptor MAVS via the HECT ubiquitin ligase AIP4. Nat Immunol. 2009;10(12):1300–8. https://doi.org/10.1038/ni.1815.
Gao P, Ma X, Yuan M, Yi Y, Liu G, Wen M, et al. E3 ligase Nedd4l promotes antiviral innate immunity by catalyzing K29-linked cysteine ubiquitination of TRAF3. Nat Commun. 2021;12(1):1194. https://doi.org/10.1038/s41467-021-21456-1.
Yuan C, Qi J, Zhao X, Gao C. Smurf1 protein negatively regulates interferon-γ signaling through promoting STAT1 protein ubiquitination and degradation. J Biol Chem. 2012;287(21):17006–15. https://doi.org/10.1074/jbc.M112.341198.
Li S, Lu K, Wang J, An L, Yang G, Chen H, et al. Ubiquitin ligase Smurf1 targets TRAF family proteins for ubiquitination and degradation. Mol Cell Biochem. 2010;338(1–2):11–7. https://doi.org/10.1007/s11010-009-0315-y.
Peng Z, Yue Y, **ong S. Mycobacterial PPE36 Modulates Host Inflammation by Promoting E3 Ligase Smurf1-Mediated MyD88 Degradation. Front Immunol. 2022;13:690667. https://doi.org/10.3389/fimmu.2022.690667.
Lee YS, Park JS, Kim JH, Jung SM, Lee JY, Kim SJ, et al. Smad6-specific recruitment of Smurf E3 ligases mediates TGF-β1-induced degradation of MyD88 in TLR4 signalling. Nat Commun. 2011;2:460. https://doi.org/10.1038/ncomms1469.
Pan Y, Li R, Meng JL, Mao HT, Zhang Y, Zhang J. Smurf2 negatively modulates RIG-I-dependent antiviral response by targeting VISA/MAVS for ubiquitination and degradation. J Immunol (Baltimore, Md : 1950). 2014;192(10):4758–64. https://doi.org/10.4049/jimmunol.1302632.
Yang Y, Liao B, Wang S, Yan B, ** Y, Shu HB, et al. E3 ligase WWP2 negatively regulates TLR3-mediated innate immune response by targeting TRIF for ubiquitination and degradation. Proc Natl Acad Sci U S A. 2013;110(13):5115–20. https://doi.org/10.1073/pnas.1220271110.
Meroni G, Diez-Roux G. TRIM/RBCC, a novel class of “single protein RING finger” E3 ubiquitin ligases. BioEssays : News Rev Mole Cell Dev Biol. 2005;27(11):1147–57. https://doi.org/10.1002/bies.20304.
Sardiello M, Cairo S, Fontanella B, Ballabio A, Meroni G. Genomic analysis of the TRIM family reveals two groups of genes with distinct evolutionary properties. BMC Evol Biol. 2008;8:225. https://doi.org/10.1186/1471-2148-8-225.
Deshaies RJ, Joazeiro CA. RING domain E3 ubiquitin ligases. Ann Rev Biochem. 2009;78:399–434. https://doi.org/10.1146/annurev.biochem.78.101807.093809.
Eisenhaber B, Chumak N, Eisenhaber F, Hauser MT. The ring between ring fingers (RBR) protein family. Genome Biol. 2007;8(3):209. https://doi.org/10.1186/gb-2007-8-3-209.
Marín I, Ferrús A. Comparative genomics of the RBR family, including the Parkinson’s disease-related gene parkin and the genes of the ariadne subfamily. Mol Biol Evol. 2002;19(12):2039–50. https://doi.org/10.1093/oxfordjournals.molbev.a004029.
Shi M, Cho H, Inn KS, Yang A, Zhao Z, Liang Q, et al. Negative regulation of NF-κB activity by brain-specific TRIpartite Motif protein 9. Nat Commun. 2014;5:4820. https://doi.org/10.1038/ncomms5820.
Qin Y, Liu Q, Tian S, **e W, Cui J, Wang RF. TRIM9 short isoform preferentially promotes DNA and RNA virus-induced production of type I interferon by recruiting GSK3β to TBK1. Cell Res. 2016;26(5):613–28. https://doi.org/10.1038/cr.2016.27.
Zhu Q, Yu T, Gan S, Wang Y, Pei Y, Zhao Q, et al. TRIM24 facilitates antiviral immunity through mediating K63-linked TRAF3 ubiquitination. J Exp Med. 2020;217:7. https://doi.org/10.1084/jem.20192083.
Martín-Vicente M, Medrano LM, Resino S, García-Sastre A, Martínez I. TRIM25 in the Regulation of the Antiviral Innate Immunity. Front Immunol. 2017;8:1187. https://doi.org/10.3389/fimmu.2017.01187.
Zhao J, Cai B, Shao Z, Zhang L, Zheng Y, Ma C, et al. TRIM26 positively regulates the inflammatory immune response through K11-linked ubiquitination of TAB1. Cell Death Differ. 2021;28(11):3077–91. https://doi.org/10.1038/s41418-021-00803-1.
Liu ZS, Zhang ZY, Cai H, Zhao M, Mao J, Dai J, et al. RINCK-mediated monoubiquitination of cGAS promotes antiviral innate immune responses. Cell & Biosci. 2018;8:35. https://doi.org/10.1186/s13578-018-0233-3.
Seo GJ, Kim C, Shin WJ, Sklan EH, Eoh H, Jung JU. TRIM56-mediated monoubiquitination of cGAS for cytosolic DNA sensing. Nat Commun. 2018;9(1):613. https://doi.org/10.1038/s41467-018-02936-3.
Liu Y, Chen Y, Ding C, Zhu X, Song X, Ren Y, et al. TRIM56 positively regulates TNFα-induced NF-κB signaling by enhancing the ubiquitination of TAK1. Int J Biol Macromol. 2022;219:571–8. https://doi.org/10.1016/j.ijbiomac.2022.08.019.
Wang Q, Huang L, Hong Z, Lv Z, Mao Z, Tang Y, et al. The E3 ubiquitin ligase RNF185 facilitates the cGAS-mediated innate immune response. PLoS Pathogens. 2017;13(3):e1006264. https://doi.org/10.1371/journal.ppat.1006264.
Yang X, Shi C, Li H, Shen S, Su C, Yin H. MARCH8 attenuates cGAS-mediated innate immune responses through ubiquitylation. Sci Signal. 2022;15(732):eabk3067. https://doi.org/10.1126/scisignal.abk3067.
Kong Z, Yin H, Wang F, Liu Z, Luan X, Sun L, et al. Pseudorabies virus tegument protein UL13 recruits RNF5 to inhibit STING-mediated antiviral immunity. PLoS Pathogens. 2022;18(5):e1010544. https://doi.org/10.1371/journal.ppat.1010544.
Wang Y, Cui S, **n T, Wang X, Yu H, Chen S, et al. African Swine Fever Virus MGF360–14L Negatively Regulates Type I Interferon Signaling by Targeting IRF3. Front Cell Infect Microbiol. 2021;11:818969. https://doi.org/10.3389/fcimb.2021.818969.
Huang L, Xu W, Liu H, Xue M, Liu X, Zhang K, et al. African Swine Fever Virus pI215L Negatively Regulates cGAS-STING Signaling Pathway through Recruiting RNF138 to Inhibit K63-Linked Ubiquitination of TBK1. J Immunol (Baltimore, Md : 1950). 2021;207(11):2754–69. https://doi.org/10.4049/jimmunol.2100320.
Suresh HG, Pascoe N, Andrews B. The structure and function of deubiquitinases: lessons from budding yeast. Open Biol. 2020;10(10):200279. https://doi.org/10.1098/rsob.200279.
Komander D, Clague MJ, Urbé S. Breaking the chains: structure and function of the deubiquitinases. Nat Rev Mol Cell Biol. 2009;10(8):550–63. https://doi.org/10.1038/nrm2731.
Engström O, Belda O, Kullman-Magnusson M, Rapp M, Böhm K, Paul R, et al. Discovery of USP7 small-molecule allosteric inhibitors. Bioorg Med Chem Lett. 2020;30(20):127471. https://doi.org/10.1016/j.bmcl.2020.127471.
Hu M, Li P, Li M, Li W, Yao T, Wu JW, et al. Crystal structure of a UBP-family deubiquitinating enzyme in isolation and in complex with ubiquitin aldehyde. Cell. 2002;111(7):1041–54. https://doi.org/10.1016/s0092-8674(02)01199-6.
Baek SH, Park KC, Lee JI, Kim KI, Yoo YJ, Tanaka K, et al. A novel family of ubiquitin-specific proteases in chick skeletal muscle with distinct N- and C-terminal extensions. Biochem J. 1998;334(Pt 3):677–84. https://doi.org/10.1042/bj3340677.
Zhang L, Zhao X, Zhang M, Zhao W, Gao C. Ubiquitin-specific protease 2b negatively regulates IFN-β production and antiviral activity by targeting TANK-binding kinase 1. J Immunol (Baltimore, Md : 1950). 2014;193(5):2230–7. https://doi.org/10.4049/jimmunol.1302634.
Zhou Q, **ao Z, Zhou R, Zhou Y, Fu P, Li X, et al. Ubiquitin-specific protease 3 targets TRAF6 for deubiquitination and suppresses IL-1β induced chondrocyte apoptosis. Biochem Biophys Res Commun. 2019;514(2):482–9. https://doi.org/10.1016/j.bbrc.2019.04.163.
Cui J, Song Y, Li Y, Zhu Q, Tan P, Qin Y, et al. USP3 inhibits type I interferon signaling by deubiquitinating RIG-I-like receptors. Cell Res. 2014;24(4):400–16. https://doi.org/10.1038/cr.2013.170.
Zhou F, Zhang X, van Dam H, Ten Dijke P, Huang H, Zhang L. Ubiquitin-specific protease 4 mitigates Toll-like/interleukin-1 receptor signaling and regulates innate immune activation. J Biol Chem. 2012;287(14):11002–10. https://doi.org/10.1074/jbc.M111.328187.
Colleran A, Collins PE, O’Carroll C, Ahmed A, Mao X, McManus B, et al. Deubiquitination of NF-κB by Ubiquitin-Specific Protease-7 promotes transcription. Proc Natl Acad Sci U S A. 2013;110(2):618–23. https://doi.org/10.1073/pnas.1208446110.
Mitxitorena I, Somma D, Mitchell JP, Lepistö M, Tyrchan C, Smith EL, et al. The deubiquitinase USP7 uses a distinct ubiquitin-like domain to deubiquitinate NF-ĸB subunits. J Biol Chem. 2020;295(33):11754–63. https://doi.org/10.1074/jbc.RA120.014113.
Zhang Y, Luo Y, Wang Y, Liu H, Yang Y, Wang Q. Effect of deubiquitinase USP8 on hypoxia/reoxygenation-induced inflammation by deubiquitination of TAK1 in renal tubular epithelial cells. Int J Mol Med. 2018;42(6):3467–76. https://doi.org/10.3892/ijmm.2018.3881.
Jiangqiao Z, Tianyu W, Zhongbao C, Long Z, Jilin Z, **aoxiong M, et al. Ubiquitin-Specific Peptidase 10 Protects Against Hepatic Ischaemic/Reperfusion Injury via TAK1 Signalling. Front Immunol. 2020;11:506275. https://doi.org/10.3389/fimmu.2020.506275.
Wang L, Wu D, Xu Z. USP10 protects against cerebral ischemia injury by suppressing inflammation and apoptosis through the inhibition of TAK1 signaling. Biochem Biophys Res Commun. 2019;516(4):1272–8. https://doi.org/10.1016/j.bbrc.2019.06.042.
Li H, Quan J, Zhao X, Ling J, Chen W. USP14 negatively regulates RIG-I-mediated IL-6 and TNF-α production by inhibiting NF-κB activation. Mole Immunol. 2021;130:69–76. https://doi.org/10.1016/j.molimm.2020.12.022.
Hou J, Han L, Zhao Z, Liu H, Zhang L, Ma C, et al. USP18 positively regulates innate antiviral immunity by promoting K63-linked polyubiquitination of MAVS. Nat Commun. 2021;12(1):2970. https://doi.org/10.1038/s41467-021-23219-4.
Hu B, Ge C, Zhu C. USP18 negatively regulates and inhibits lipopolysaccharides-induced sepsis by targeting TAK1 activity. Int Immunol. 2021. https://doi.org/10.1093/intimm/dxab029.
Yang Z, **an H, Hu J, Tian S, Qin Y, Wang RF, et al. USP18 negatively regulates NF-κB signaling by targeting TAK1 and NEMO for deubiquitination through distinct mechanisms. Sci Rep. 2015;5:12738. https://doi.org/10.1038/srep12738.
Lei CQ, Wu X, Zhong X, Jiang L, Zhong B, Shu HB. USP19 Inhibits TNF-α- and IL-1β-Triggered NF-κB Activation by Deubiquitinating TAK1. J Immunol (Baltimore, Md : 1950). 2019;203(1):259–68. https://doi.org/10.4049/jimmunol.1900083.
Gu Z, Shi W, Zhang L, Hu Z, Xu C. USP19 suppresses cellular type I interferon signaling by targeting TRAF3 for deubiquitination. Future Microbiol. 2017;12:767–79. https://doi.org/10.2217/fmb-2017-0006.
Miao R, Lu Y, He X, Liu X, Chen Z, Wang J. Ubiquitin-specific protease 19 blunts pathological cardiac hypertrophy via inhibition of the TAK1-dependent pathway. J Cell Mol Med. 2020;24(18):10946–57. https://doi.org/10.1111/jcmm.15724.
Jean-Charles PY, Wu JH, Zhang L, Kaur S, Nepliouev I, Stiber JA, et al. USP20 (Ubiquitin-Specific Protease 20) Inhibits TNF (Tumor Necrosis Factor)-Triggered Smooth Muscle Cell Inflammation and Attenuates Atherosclerosis. Arterioscler Thromb Vasc Biol. 2018;38(10):2295–305. https://doi.org/10.1161/atvbaha.118.311071.
Fan Y, Mao R, Yu Y, Liu S, Shi Z, Cheng J, et al. USP21 negatively regulates antiviral response by acting as a RIG-I deubiquitinase. J Exp Med. 2014;211(2):313–28. https://doi.org/10.1084/jem.20122844.
Lin D, Zhang M, Zhang MX, Ren Y, ** J, Zhao Q, et al. Induction of USP25 by viral infection promotes innate antiviral responses by mediating the stabilization of TRAF3 and TRAF6. Proc Natl Acad Sci U S A. 2015;112(36):11324–9. https://doi.org/10.1073/pnas.1509968112.
Zhong B, Liu X, Wang X, Liu X, Li H, Darnay BG, et al. Ubiquitin-specific protease 25 regulates TLR4-dependent innate immune responses through deubiquitination of the adaptor protein TRAF3. Sci Signal. 2013;6(275):ra35. https://doi.org/10.1126/scisignal.2003708.
Zhong B, Liu X, Wang X, Chang SH, Liu X, Wang A, et al. Negative regulation of IL-17-mediated signaling and inflammation by the ubiquitin-specific protease USP25. Nat Immunol. 2012;13(11):1110–7. https://doi.org/10.1038/ni.2427.
Guo Y, Jiang F, Kong L, Li B, Yang Y, Zhang L, et al. Cutting Edge: USP27X Deubiquitinates and Stabilizes the DNA Sensor cGAS to Regulate Cytosolic DNA-Mediated Signaling. J Immunol (Baltimore, Md : 1950). 2019;203(8):2049–54. https://doi.org/10.4049/jimmunol.1900514.
Lin M, Zhao Z, Yang Z, Meng Q, Tan P, **e W, et al. USP38 Inhibits Type I Interferon Signaling by Editing TBK1 Ubiquitination through NLRP4 Signalosome. Mol Cell. 2016;64(2):267–81. https://doi.org/10.1016/j.molcel.2016.08.029.
Peng Y, Guo J, Sun T, Fu Y, Zheng H, Dong C, et al. USP39 Serves as a Deubiquitinase to Stabilize STAT1 and Sustains Type I IFN-Induced Antiviral Immunity. J Immunol (Baltimore, Md : 1950). 2020;205(11):3167–78. https://doi.org/10.4049/jimmunol.1901384.
Lei H, Yang L, Xu H, Wang Z, Li X, Liu M, et al. Ubiquitin-specific protease 47 regulates intestinal inflammation through deubiquitination of TRAF6 in epithelial cells. Sci China Life Sci. 2022. https://doi.org/10.1007/s11427-021-2040-8.
Du J, Fu L, Sui Y, Zhang L. The function and regulation of OTU deubiquitinases. Front Med. 2020;14(5):542–63. https://doi.org/10.1007/s11684-019-0734-4.
Zhang L, Liu J, Qian L, Feng Q, Wang X, Yuan Y, et al. Induction of OTUD1 by RNA viruses potently inhibits innate immune responses by promoting degradation of the MAVS/TRAF3/TRAF6 signalosome. PLoS Pathogens. 2018;14(5):e1007067. https://doi.org/10.1371/journal.ppat.1007067.
Wu B, Qiang L, Zhang Y, Fu Y, Zhao M, Lei Z, et al. The deubiquitinase OTUD1 inhibits colonic inflammation by suppressing RIPK1-mediated NF-κB signaling. Cell Mol Immunol. 2022;19(2):276–89. https://doi.org/10.1038/s41423-021-00810-9.
Zhang Z, Fang X, Wu X, Ling L, Chu F, Li J, et al. Acetylation-Dependent Deubiquitinase OTUD3 Controls MAVS Activation in Innate Antiviral Immunity. Mol Cell. 2020;79(2):304-19.e7. https://doi.org/10.1016/j.molcel.2020.06.020.
Zhao Y, Mudge MC, Soll JM, Rodrigues RB, Byrum AK, Schwarzkopf EA, et al. OTUD4 Is a Phospho-Activated K63 Deubiquitinase that Regulates MyD88-Dependent Signaling. Mol Cell. 2018;69(3):505-16.e5. https://doi.org/10.1016/j.molcel.2018.01.009.
Liuyu T, Yu K, Ye L, Zhang Z, Zhang M, Ren Y, et al. Induction of OTUD4 by viral infection promotes antiviral responses through deubiquitinating and stabilizing MAVS. Cell Res. 2019;29(1):67–79. https://doi.org/10.1038/s41422-018-0107-6.
Liu H, Fan J, Zhang W, Chen Q, Zhang Y, Wu Z. OTUD4 alleviates hepatic ischemia-reperfusion injury by suppressing the K63-linked ubiquitination of TRAF6. Biochem Biophys Res Commun. 2020;523(4):924–30. https://doi.org/10.1016/j.bbrc.2019.12.114.
Guo Y, Jiang F, Kong L, Wu H, Zhang H, Chen X, et al. OTUD5 promotes innate antiviral and antitumor immunity through deubiquitinating and stabilizing STING. Cell Mol Immunol. 2021;18(8):1945–55. https://doi.org/10.1038/s41423-020-00531-5.
Zhou Z, Cai X, Zhu J, Li Z, Yu G, Liu X, et al. Zebrafish otud6b Negatively Regulates Antiviral Responses by Suppressing K63-Linked Ubiquitination of irf3 and irf7. J Immunol (Baltimore, Md : 1950). 2021;207(1):244–56. https://doi.org/10.4049/jimmunol.2000891.
**e W, Tian S, Yang J, Cai S, ** S, Zhou T, et al. OTUD7B deubiquitinates SQSTM1/p62 and promotes IRF3 degradation to regulate antiviral immunity. Autophagy. 2022;18(10):2288–302. https://doi.org/10.1080/15548627.2022.2026098.
Li Y, Mooney EC, Holden SE, **a XJ, Cohen DJ, Walsh SW, et al. A20 Orchestrates Inflammatory Response in the Oral Mucosa through Restraining NF-κB Activity. J Immunol (Baltimore, Md : 1950). 2019;202(7):2044–56. https://doi.org/10.4049/jimmunol.1801286.
Jang JH, Kim H, Jung IY, Cho JH. A20 Inhibits LPS-Induced Inflammation by Regulating TRAF6 Polyubiquitination in Rainbow Trout. Int J Mol Sci. 2021;22:18. https://doi.org/10.3390/ijms22189801.
Mooney EC, Sahingur SE. The Ubiquitin System and A20: Implications in Health and Disease. J Dent Res. 2021;100(1):10–20. https://doi.org/10.1177/0022034520949486.
Arguello M, Paz S, Ferran C, Moll HP, Hiscott J. Anti-viral tetris: modulation of the innate anti-viral immune response by A20. Adv Exp Med Biol. 2014;809:49–64. https://doi.org/10.1007/978-1-4939-0398-6_4.
Licchesi JD, Mieszczanek J, Mevissen TE, Rutherford TJ, Akutsu M, Virdee S, et al. An ankyrin-repeat ubiquitin-binding domain determines TRABID’s specificity for atypical ubiquitin chains. Nat Struct Mol Biol. 2011;19(1):62–71. https://doi.org/10.1038/nsmb.2169.
** J, **e X, **ao Y, Hu H, Zou Q, Cheng X, et al. Epigenetic regulation of the expression of Il12 and Il23 and autoimmune inflammation by the deubiquitinase Trabid. Nat Immunol. 2016;17(3):259–68. https://doi.org/10.1038/ni.3347.
Keusekotten K, Elliott PR, Glockner L, Fiil BK, Damgaard RB, Kulathu Y, et al. OTULIN antagonizes LUBAC signaling by specifically hydrolyzing Met1-linked polyubiquitin. Cell. 2013;153(6):1312–26. https://doi.org/10.1016/j.cell.2013.05.014.
Rivkin E, Almeida SM, Ceccarelli DF, Juang YC, MacLean TA, Srikumar T, et al. The linear ubiquitin-specific deubiquitinase gumby regulates angiogenesis. Nature. 2013;498(7454):318–24. https://doi.org/10.1038/nature12296.
Elliott PR, Nielsen SV, Marco-Casanova P, Fiil BK, Keusekotten K, Mailand N, et al. Molecular basis and regulation of OTULIN-LUBAC interaction. Mol Cell. 2014;54(3):335–48. https://doi.org/10.1016/j.molcel.2014.03.018.
Damgaard RB, Walker JA, Marco-Casanova P, Morgan NV, Titheradge HL, Elliott PR, et al. The Deubiquitinase OTULIN Is an Essential Negative Regulator of Inflammation and Autoimmunity. Cell. 2016;166(5):1215-30.e20. https://doi.org/10.1016/j.cell.2016.07.019.
Heger K, Wickliffe KE, Ndoja A, Zhang J, Murthy A, Dugger DL, et al. OTULIN limits cell death and inflammation by deubiquitinating LUBAC. Nature. 2018;559(7712):120–4. https://doi.org/10.1038/s41586-018-0256-2.
Buckley SM, Aranda-Orgilles B, Strikoudis A, Apostolou E, Loizou E, Moran-Crusio K, et al. Regulation of pluripotency and cellular reprogramming by the ubiquitin-proteasome system. Cell Stem Cell. 2012;11(6):783–98. https://doi.org/10.1016/j.stem.2012.09.011.
Butler LR, Densham RM, Jia J, Garvin AJ, Stone HR, Shah V, et al. The proteasomal de-ubiquitinating enzyme POH1 promotes the double-strand DNA break response. EMBO J. 2012;31(19):3918–34. https://doi.org/10.1038/emboj.2012.232.
Cooper EM, Cutcliffe C, Kristiansen TZ, Pandey A, Pickart CM, Cohen RE. K63-specific deubiquitination by two JAMM/MPN+ complexes: BRISC-associated Brcc36 and proteasomal Poh1. EMBO J. 2009;28(6):621–31. https://doi.org/10.1038/emboj.2009.27.
Wang B, Ma A, Zhang L, ** WL, Qian Y, Xu G, et al. POH1 deubiquitylates and stabilizes E2F1 to promote tumour formation. Nat Commun. 2015;6:8704. https://doi.org/10.1038/ncomms9704.
Py BF, Kim MS, Vakifahmetoglu-Norberg H, Yuan J. Deubiquitination of NLRP3 by BRCC3 critically regulates inflammasome activity. Mol Cell. 2013;49(2):331–8. https://doi.org/10.1016/j.molcel.2012.11.009.
Bednash JS, Johns F, Patel N, Smail TR, Londino JD, Mallampalli RK. The deubiquitinase STAMBP modulates cytokine secretion through the NLRP3 inflammasome. Cell Signal. 2021;79:109859. https://doi.org/10.1016/j.cellsig.2020.109859.
Bednash JS, Weathington N, Londino J, Rojas M, Gulick DL, Fort R, et al. Targeting the deubiquitinase STAMBP inhibits NALP7 inflammasome activity. Nat Commun. 2017;8:15203. https://doi.org/10.1038/ncomms15203.
Tian M, Liu W, Zhang Q, Huang Y, Li W, Wang W, et al. MYSM1 Represses Innate Immunity and Autoimmunity through Suppressing the cGAS-STING Pathway. Cell Reports. 2020;33(3):108297. https://doi.org/10.1016/j.celrep.2020.108297.
Panda S, Nilsson JA, Gekara NO. Deubiquitinase MYSM1 Regulates Innate Immunity through Inactivation of TRAF3 and TRAF6 Complexes. Immunity. 2015;43(4):647–59. https://doi.org/10.1016/j.immuni.2015.09.010.
Zhang L, Wei N, Cui Y, Hong Z, Liu X, Wang Q, et al. The deubiquitinase CYLD is a specific checkpoint of the STING antiviral signaling pathway. PLoS pathogens. 2018;14(11):e1007435. https://doi.org/10.1371/journal.ppat.1007435.
Abdul Rehman SA, Kristariyanto YA, Choi SY, Nkosi PJ, Weidlich S, Labib K, et al. MINDY-1 Is a Member of an Evolutionarily Conserved and Structurally Distinct New Family of Deubiquitinating Enzymes. Mol Cell. 2016;63(1):146–55. https://doi.org/10.1016/j.molcel.2016.05.009.
Maurer T, Wertz IE. Length Matters: MINDY Is a New Deubiquitinase Family that Preferentially Cleaves Long Polyubiquitin Chains. Mol Cell. 2016;63(1):4–6. https://doi.org/10.1016/j.molcel.2016.06.027.
Tang J, Luo Y, Long G, Zhou L. MINDY1 promotes breast cancer cell proliferation by stabilizing estrogen receptor α. Cell Death & Dis. 2021;12(10):937. https://doi.org/10.1038/s41419-021-04244-z.
Wang S, Wang K, Li J, Zheng C. Herpes simplex virus 1 ubiquitin-specific protease UL36 inhibits beta interferon production by deubiquitinating TRAF3. J Virol. 2013;87(21):11851–60. https://doi.org/10.1128/jvi.01211-13.
Ye R, Su C, Xu H, Zheng C. Herpes Simplex Virus 1 Ubiquitin-Specific Protease UL36 Abrogates NF-κB Activation in DNA Sensing Signal Pathway. J Virol. 2017;91:5. https://doi.org/10.1128/jvi.02417-16.
**ao Y, Huang Q, Wu Z, Chen W. Roles of protein ubiquitination in inflammatory bowel disease. Immunobiology. 2020;225(6):152026. https://doi.org/10.1016/j.imbio.2020.152026.
Funding
This study was supported by grants from The Natural Science Foundation of Hunan Province (No.2023JJ30067).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Jiang, W., Li, M., Peng, S. et al. Ubiquitin ligase enzymes and de-ubiquitinating enzymes regulate innate immunity in the TLR, NLR, RLR, and cGAS-STING pathways. Immunol Res 71, 800–813 (2023). https://doi.org/10.1007/s12026-023-09400-5
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12026-023-09400-5