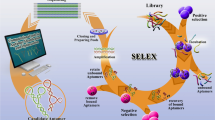
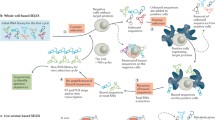
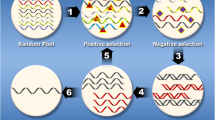
Abstract
Conventional cancer therapies can have significant adverse effects as they are not targeted to cancer cells and may damage healthy cells. Single-stranded oligonucleotides assembled in a particular architecture, known as aptamers, enable them to attach selectively to target areas. Usually, they are created by Systematic Evolution of Ligand by Exponential enrichment (SELEX), and they go through a rigorous pharmacological revision process to change their therapeutic half-life, affinity, and specificity. They could thus offer a viable substitute for antibodies in the targeted cancer treatment market. Although aptamers can be a better choice in some situations, antibodies are still appropriate for many other uses. The technique of delivering aptamers is simple and reasonable, and the time needed to manufacture them is relatively brief. Aptamers do not require animals or an immune response to be produced, in contrast to antibodies. When used as a medication, aptamers can directly suppress tumor cells. As an alternative, they can be included in systems for targeted drug delivery that administer medications specifically to tumor cells while reducing toxicity to healthy cells. The most recent and cutting-edge methods for treating gastrointestinal (GI) tract cancer with aptamers will be covered in this review, with a focus on targeted therapy as a means of conquering resistance to traditional medicines.

Similar content being viewed by others
References
Adachi, T. & Nakamura, Y. (2019). Aptamers: A Review of Their Chemical Properties and Modifications for Therapeutic Application. Molecules (Basel, Switzerland), 24, 29–42.
Byun, J. (2021). Recent Progress and Opportunities for Nucleic Acid Aptamers. Life (Basel, Switzerland), 11, 193.
Yang, Y., Ren, X., Schluesener, H. J., & Zhang, Z. (2011). Aptamers: selection, modification and application to nervous system diseases. Current medicinal chemistry, 18, 4159–4168.
Yazdian-Robati, R., Arab, A., Ramezani, M., Abnous, K., & Taghdisi, S. M. (2017). Application of aptamers in treatment and diagnosis of leukemia. International journal of pharmaceutics, 529, 44–54.
Emami, N., Pakchin, P. S., & Ferdousi, R. (2020). Computational predictive approaches for interaction and structure of aptamers. Journal of theoretical biology, 497, 110268.
Zhu, G., & Chen, X. (2018). Aptamer-based targeted therapy. Advanced drug delivery reviews, 134, 65–78.
Kanwar, J. R., Roy, K., & Kanwar, R. K. (2011). Chimeric aptamers in cancer cell-targeted drug delivery. Critical reviews in biochemistry and molecular biology, 46, 459–477.
Kinghorn, A. B., Fraser, L. A., Lang, S., Shiu, S. C. C. & Tanner, J. A. (2017). Aptamer Bioinformatics. International journal of molecular sciences, 18, 2516.
Kumar Kulabhusan, P., Hussain, B. & Yüce, M. (2020). Current Perspectives on Aptamers as Diagnostic Tools and Therapeutic Agents. Pharmaceutics, 12, 646
Yan, A. C., & Levy, M. (2009). Aptamers and aptamer targeted delivery. RNA biology, 6, 316–320.
Nimjee, S. M., White, R. R., Becker, R. C., & Sullenger, B. A. (2017). Aptamers as Therapeutics. Annual review of pharmacology and toxicology, 57, 61–79.
Radom, F., Jurek, P. M., Mazurek, M. P., Otlewski, J., & Jeleń, F. (2013). Aptamers: molecules of great potential. Biotechnology advances, 31, 1260–1274.
Tan, Y., Ma, L., Yang, X., Cheng, Q. N., & Wu, J. F. (2023). Current Status and Challenges of Aptamers Screening and Optimization. Combinatorial chemistry & high throughput screening., 26, 1067–1082.
Wu, Y. X., & Kwon, Y. J. (2016). Aptamers: The “evolution” of SELEX. Methods (San Diego, Calif), 106, 21–28.
Sullenger, B. A. (2016). Aptamers Coming of Age at Twenty-Five. Nucleic acid therapeutics, 26, 119.
Xu, Y., Yang, X., & Wang, E. (2010). Review: Aptamers in microfluidic chips. Analytica chimica acta, 683, 12–20.
Rozenblum, G. T., Lopez, V. G., Vitullo, A. D., & Radrizzani, M. (2016). Aptamers: current challenges and future prospects. Expert opinion on drug discovery, 11, 127–135.
Soldevilla, M. M., Hervas, S., Villanueva, H., Lozano, T., Rabal, O., & Oyarzabal, J., et al. (2017). Identification of LAG3 high affinity aptamers by HT-SELEX and Conserved Motif Accumulation (CMA). PloS one, 12, e0185169.
Saad, M., & Faucher, S. P. (2021). Aptamers and Aptamer-Coupled Biosensors to Detect Water-Borne Pathogens. Frontiers in microbiology, 12, 643797.
Schmitz, F. R. W., Valério, A., de Oliveira, D., & Hotza, D. (2020). An overview and future prospects on aptamers for food safety. Applied microbiology and biotechnology, 104, 6929–6939.
Santulli-Marotto, S., Nair, S. K., Rusconi, C., Sullenger, B. & Gilboa, EJCR. (2003). Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer Research, 63, 7483–7489.
Zhou, B., Mo, Z., Lai, G., Chen, X., Li, R., & Wu, R., et al. (2023). Targeting tumor exosomal circular RNA cSERPINE2 suppresses breast cancer progression by modulating MALT1-NF-. Journal of experimental & clinical cancer research: CR, 42, 48.
Prodeus, A., Abdul-Wahid, A., Fischer, N. W., Huang, E. H., Cydzik, M., & Gariépy, J. (2015). Targeting the PD-1/PD-L1 Immune Evasion Axis With DNA Aptamers as a Novel Therapeutic Strategy for the Treatment of Disseminated Cancers. Molecular therapy Nucleic acids, 4, e237.
Gefen, T., Castro, I., Muharemagic, D., Puplampu-Dove, Y., Patel, S., & Gilboa, E. (2017). A TIM-3 Oligonucleotide Aptamer Enhances T Cell Functions and Potentiates Tumor Immunity in Mice. Molecular therapy: the journal of the American Society of Gene Therapy, 25, 2280–2288.
Gilboa, E., McNamara, 2nd, J., & Pastor, F. (2013). Use of oligonucleotide aptamer ligands to modulate the function of immune receptors. Clinical cancer research: an official journal of the American Association for Cancer Research, 19, 1054–1062.
Zhou, G., Wilson, G., Hebbard, L., Duan, W., Liddle, C., & George, J., et al. (2016). Aptamers: A promising chemical antibody for cancer therapy. Oncotarget, 7, 13446–13463.
Pratico, E. D., Sullenger, B. A., & Nair, S. K. (2013). Identification and characterization of an agonistic aptamer against the T cell costimulatory receptor, OX40. Nucleic acid therapeutics, 23, 35–43.
Pastor, F., Soldevilla, M. M., Villanueva, H., Kolonias, D., Inoges, S., & de Cerio, A. L., et al. (2013). CD28 aptamers as powerful immune response modulators. Molecular therapy Nucleic acids, 2, e98.
Massari, F., Santoni, M., Ciccarese, C., Santini, D., Alfieri, S., & Martignoni, G., et al. (2015). PD-1 blockade therapy in renal cell carcinoma: current studies and future promises. Cancer treatment reviews, 41, 114–121.
Eder, J. P., Vande Woude, G. F., Boerner, S. A., & LoRusso, P. M. (2009). Novel therapeutic inhibitors of the c-Met signaling pathway in cancer. Clinical cancer research: an official journal of the American Association for Cancer Research, 15, 2207–2214.
Borghaei, H., Paz-Ares, L., Horn, L., Spigel, D. R., Steins, M., & Ready, N. E., et al. (2015). Nivolumab versus Docetaxel in Advanced Nonsquamous Non-Small-Cell Lung Cancer. The New England journal of medicine, 373, 1627–1639.
Goh, K. W., Stephen, A., Wu, Y. S., Sim, M. S., Batumalaie, K., & Gopinath, S. C. B., et al. (2023). Molecular Targets of Aptamers in Gastrointestinal Cancers: Cancer Detection, Therapeutic Applications, and Associated Mechanisms. J Cancer, 14, 2491–2516.
Chen, A., & Yang, S. (2015). Replacing antibodies with aptamers in lateral flow immunoassay. Biosensors & bioelectronics, 71, 230–242.
Bayat, P., Nosrati, R., Alibolandi, M., Rafatpanah, H., Abnous, K., & Khedri, M., et al. (2018). SELEX methods on the road to protein targeting with nucleic acid aptamers. Biochimie, 154, 132–155.
Marrazza G. (2017). Aptamer Sensors. Biosensors.7,5.
Mattice, C. M., & DeRosa, M. C. (2015). Status and Prospects of Aptamers as Drug Components. BioDrugs: clinical immunotherapeutics, biopharmaceuticals and gene therapy, 29, 151–165.
Wu, L., Wang, Y., Xu, X., Liu, Y., Lin, B., & Zhang, M., et al. (2021). Aptamer-Based Detection of Circulating Targets for Precision Medicine. Chemical reviews, 121, 12035–12105.
Gu, L., Zheng, J., Zhang, Y., Wang, D., & Liu, J. (2023). Capture-SELEX of DNA Aptamers for Sulforhodamine B and Fluorescein. Chemistry (Weinheim an der Bergstrasse, Germany), 29, e202302616.
Ducongé, F. (2023). Improvement of Aptamers by High-Throughput Sequencing of Doped-SELEX. Methods in molecular biology (Clifton, NJ), 2570, 85–102.
Dua, P., Kim, S., & Lee, D. K. (2008). Patents on SELEX and therapeutic aptamers. Recent patents on DNA & gene sequences., 2, 172–186.
Gao, S., Zheng, X., Jiao, B., & Wang, L. (2016). Post-SELEX optimization of aptamers. Analytical and bioanalytical chemistry, 408, 4567–4573.
Zhong, Y., Zhao, J., Li, J., Liao, X., & Chen, F. (2020). Advances of aptamers screened by Cell-SELEX in selection procedure, cancer diagnostics and therapeutics. Analytical biochemistry, 598, 113620.
Zhu, C., Feng, Z., Qin, H., Chen, L., Yan, M., & Li, L., et al. (2024). Recent progress of SELEX methods for screening nucleic acid aptamers. Talanta, 266, 124998.
Kohlberger, M., & Gadermaier, G. (2022). SELEX: Critical factors and optimization strategies for successful aptamer selection. Biotechnology and applied biochemistry, 69, 1771–1792.
Ye, M., Hu, J., Peng, M., Liu, J., Liu, J., & Liu, H., et al. (2012). Generating aptamers by cell-SELEX for applications in molecular medicine. International journal of molecular sciences, 13, 3341–3353.
Ramezanpour, M., Daei, P., Tabarzad, M., Khanaki, K., Elmi, A., & Barati, M. (2019). Preliminary study on the effect of nucleolin specific aptamer–miRNA let-7d chimera on Janus kinase-2 expression level and activity in gastric cancer (MKN-45) cells. Molecular biology reports, 46, 207–215.
Zhang, Y., Tan, J., Zhou, L., Shan, X., Liu, J., & Ma, Y. (2020). Synthesis and application of AS1411-functionalized gold nanoparticles for targeted therapy of gastric cancer. ACS omega, 5, 31227–31233.
Breitsprecher, D., Schlinck, N., Witte, D., Duhr, S., Baaske, P., & Schubert, T. (2016). Aptamer Binding Studies Using MicroScale Thermophoresis. Methods in molecular biology (Clifton, NJ), 1380, 99–111.
Minagawa, H., Onodera, K., Fujita, H., Sakamoto, T., Akitomi, J., & Kaneko, N., et al. (2017). Selection, Characterization and Application of Artificial DNA Aptamer Containing Appended Bases with Sub-nanomolar Affinity for a Salivary Biomarker. Scientific reports, 7, 42716.
Virgilio, A., Petraccone, L., Scuotto, M., Vellecco, V., Bucci, M., & Mayol, L., et al. (2014). 5-Hydroxymethyl-2’-deoxyuridine residues in the thrombin binding aptamer: investigating anticoagulant activity by making a tiny chemical modification. Chembiochem: a European journal of chemical biology, 15, 2427–2434.
Maio, G. E., Enweronye, O., Zumrut, H. E., Batool, S., Van, N. A. & Mallikaratchy, PRJC. (2017). Systematic optimization and modification of a DNA aptamer with 2'-O-methyl RNA analogues. Chemistry Select, 2, 2335–2340.
Kasahara, Y., Kitadume, S., Morihiro, K., Kuwahara, M., Ozaki, H., & Sawai, H., et al. (2010). Effect of 3′-end cap** of aptamer with various 2’,4’-bridged nucleotides: Enzymatic post-modification toward a practical use of polyclonal aptamers. Bioorganic & medicinal chemistry letters, 20, 1626–1629.
Mahmoudian, F., Ahmari, A., Shabani, S., Sadeghi, B., Fahimirad, S., & Fattahi, F. (2024). Aptamers as an approach to targeted cancer therapy. Cancer Cell Int, 24, 108.
**ang, D., Zheng, C., Zhou, S.-F., Qiao, S. & Tran, PH. et al. (2015). Superior performance of aptamer in tumor penetration over antibody: implication of aptamer-based theranostics in solid tumors. Theranostics, 5, 1083.
Chen A., Yang S. J. B. (2015). Bioelectronics. Replacing antibodies with aptamers in lateral flow immunoassay. Biosensors and bioelectronics, 71, 230–242.
Crivianu-Gaita V., Thompson M. J. B. (2016). Bioelectronics. Aptamers, antibody scFv, and antibody Fab’fragments: An overview and comparison of three of the most versatile biosensor biorecognition elements. Biosensors and Bioelectronics, 85, 32–45.
Toh S. Y., Citartan M., Gopinath S. C., Tang T-HJB. (2015). Bioelectronics. Aptamers as a replacement for antibodies in enzyme-linked immunosorbent assay. 64, 392–403.
Chen, K., Zhou, J., Shao, Z., Liu, J., Song, J. & Wang, R. et al. (2020). Aptamers as versatile molecular tools for antibody production monitoring and quality control. Journal of the American Chemical Society, 142, 12079–12086.
Li, L., Xu, S., Yan, H., Li, X., Yazd, H. S. & Li, X. et al. (2021). Nucleic acid aptamers for molecular diagnostics and therapeutics: advances and perspectives. Angewandte Chemie International Edition, 60, 2221–2231.
Han J., Gao L., Wang J., Wang JJJoC. (2020). Application and development of aptamer in cancer: from clinical diagnosis to cancer therapy. Journal of Cancer, 11, 6902.
Graham, T. A., & Sottoriva, A. (2017). Measuring cancer evolution from the genome. The Journal of pathology, 241, 183–191.
Roy, P. S., & Saikia, B. J. (2016). Cancer and cure: A critical analysis. Indian journal of cancer, 53, 441–442.
Torre, L. A., Siegel, R. L., Ward, E. M., & Jemal, A. (2016). Global Cancer Incidence and Mortality Rates and Trends–An Update. Cancer epidemiology, biomarkers & prevention : a publication of the American Association for Cancer Research, cosponsored by the American Society of Preventive Oncology, 25, 16–27.
Camorani, S., Caliendo, A., Morrone, E., Agnello, L., Martini, M., & Cantile, M., et al. (2024). Bispecific aptamer-decorated and light-triggered nanoparticles targeting tumor and stromal cells in breast cancer derived organoids: implications for precision phototherapies. Journal of experimental & clinical cancer research : CR, 43, 92.
Cunha, P. D. S., de Miranda, M. C., de Melo, M. I. A., Ferreira, A. D. F., Barbosa, J. L., & Oliveira, J. A. C., et al. (2024). Selection of internalizing RNA aptamers into human breast cancer cells derived from primary sites. Journal of cellular biochemistry, 125, e30540.
Deng, M., Yang, H., Zhang, H., Li, C., Chen, J., & Tang, W., et al. (2024). Portable and Rapid Dual-Biomarker Detection Using Solution-Gated Graphene Field Transistors in the Accurate Diagnosis of Prostate Cancer. Advanced healthcare materials, 13, e2302117.
Bahreyni, A., Mohamud, Y., Ashraf Nouhegar, S., Zhang, J., & Luo, H. (2024). Synergistic Viro-chemoimmunotherapy in Breast Cancer Enabled by Bioengineered Immunostimulatory Exosomes and Dual-Targeted Coxsackievirus B3. ACS nano, 18, 4241–4255.
Ben Moussa, F., Kutner, W., Beduk, T., Sena-Torralba, A., & Mostafavi, E. (2024). Electrochemical bio- and chemosensors for cancer biomarkers: Natural (with antibodies) versus biomimicking artificial (with aptamers and molecularly imprinted polymers) recognition. Talanta, 267, 125259.
Wen, J., Xue, L., Wei, Y., Liang, J., Jia, W., & Yong, T., et al. (2024). YTHDF2 Is a Therapeutic Target for HCC by Suppressing Immune Evasion and Angiogenesis Through ETV5/PD-L1/VEGFA Axis. Advanced science (Weinheim, Baden-Wurttemberg, Germany), 11, e2307242.
Wu, H., Lin, J., Ling, N., Zhang, Y., He, Y., & Qiu, L., et al. (2024). Functional Nucleic Acid-Based Immunomodulation for T Cell-Mediated Cancer Therapy. ACS nano, 18, 119–135.
**ang Y., Liu J., Chen J., **ao M., Pei H., Li L. (2024). MoS(2)-Based Sensor Array for Accurate Identification of Cancer Cells with Ensemble-Modified Aptamers. ACS applied materials & interfaces.16, 15861–15869.
Kianpour, M., Huang, C. W., Vejvisithsakul, P. P., Wang, J. Y., Li, C. F., & Shiao, M. S., et al. (2023). Aptamer/doxorubicin-conjugated nanoparticles target membranous CEMIP2 in colorectal cancer. International journal of biological macromolecules, 245, 125510.
Natesh, J., Chandola, C., Meeran, S. M., & Neerathilingam, M. (2021). Targeted delivery of doxorubicin through CD44 aptamer to cancer cells. Therapeutic delivery, 12, 693–703.
Nelissen, F. H. T., Peeters, W. J. M., Roelofs, T. P., Nagelkerke, A., Span, P. N., Heus, H. A. (2021). Improving Breast Cancer Treatment Specificity Using Aptamers Obtained by 3D Cell-SELEX. Pharmaceuticals (Basel, Switzerland), 14, 349.
Ding, D., Zhao, H., Wei, D., Yang, Q., Yang, C., & Wang, R., et al. (2023). The first-in-human whole-body dynamic pharmacokinetics study of Aptamer. Research, 6, 0126.
Esawi, E., Alshaer, W., Mahmoud, I. S., Alqudah, D. A., Azab, B., Awidi, A. (2021). Aptamer-Aptamer Chimera for Targeted Delivery and ATP-Responsive Release of Doxorubicin into Cancer Cells. International journal of molecular sciences, 22, 12940.
Futane, A., Jadhav, P., Mustafa, A. H., Srinivasan, A., & Narayanamurthy, V. (2024). Aptamer-functionalized MOFs and AI-driven strategies for early cancer diagnosis and therapeutics. Biotechnology letters, 46, 1–17.
Li, P., & Liu, Z. (2024). Glycan-specific molecularly imprinted polymers towards cancer diagnostics: merits, applications, and future perspectives. Chemical Society reviews, 53, 1870–1891.
Abnous, K., Danesh, N. M., Ramezani, M., Charbgoo, F., Bahreyni, A., & Taghdisi, S. M. (2018). Targeted delivery of doxorubicin to cancer cells by a cruciform DNA nanostructure composed of AS1411 and FOXM1 aptamers. Expert opinion on drug delivery, 15, 1045–1052.
Song, X., Ren, Y., Zhang, J., Wang, G., Han, X., & Zheng, W., et al. (2015). Targeted delivery of doxorubicin to breast cancer cells by aptamer functionalized DOTAP/DOPE liposomes. Oncology reports, 34, 1953–1960.
Yin, W., Pham, C. V., Wang, T., Al Shamaileh, H., Chowdhury, R., Patel S., et al. (2022). Inhibition of Autophagy Promotes the Elimination of Liver Cancer Stem Cells by CD133 Aptamer-Targeted Delivery of Doxorubicin. Biomolecules, 12, 1623.
Li, Y., Liu, W., Xu, H., Zhou, Y., **e, W., & Guo, Y., et al. (2024). Aptamers combined with immune checkpoints for cancer detection and targeted therapy: A review. International journal of biological macromolecules, 262, 130032.
Smyth, E. C., Nilsson, M., Grabsch, H. I., van Grieken, N. C., & Lordick, F. (2020). Gastric cancer. The Lancet, 396, 635–648.
Joshi, S. S., & Badgwell, B. D. (2021). Current treatment and recent progress in gastric cancer. CA: a cancer journal for clinicians, 71, 264–279.
Bie, L., Wang, Y., Jiang, F., **ao, Z., Zhang, L., & Wang, J. (2022). Insights into the binding mode of AS1411 aptamer to nucleolin. Frontiers in Molecular Biosciences, 9, 1025313.
Chen, H., Wang, J., Wang, H., Liang, J., Dong, J., & Bai, H., et al. (2022). Advances in the application of Let‑7 microRNAs in the diagnosis, treatment and prognosis of leukemia. Oncology Letters, 23, 1–8.
Wang, Y., Zhao, J., Chen, S., Li, D., Yang, J., & Zhao, X., et al. (2022). Let-7 as a promising target in aging and aging-related diseases: a promise or a pledge. Biomolecules, 12, 1070.
Yan, J., Bhadane, R., Ran, M., Ma, X., Li, Y., & Zheng, D., et al. (2024). Development of Aptamer-DNAzyme based metal-nucleic acid frameworks for gastric cancer therapy. Nature Communications, 15, 3684.
Pan, Q., Law, C. O., Yung, M. M., Han, K., Pon, Y. L., & Lau, T. C. K. (2018). Novel RNA aptamers targeting gastrointestinal cancer biomarkers CEA, CA50 and CA72-4 with superior affinity and specificity. PLoS One, 13, e0198980.
Sultana R., Badruddoza S. M., Afrin T., Sarkar A., Sarker S., Kundu R. R., et al. (2021). Serum CEA and CA 19-9 Level in Gastric Adenocarcinoma and Their Correlations with Histopathological Grading and Staging. Instruction to Authors, 40.
Wu, J. (2021). The enhanced permeability and retention (EPR) effect: the significance of the concept and methods to enhance its application. Journal of personalized medicine, 11, 771.
Angell, H. K., Bruni, D., Barrett, J. C., Herbst, R., & Galon, J. (2020). The immunoscore: colon cancer and beyond. Clinical cancer research, 26, 332–339.
Cong, Y., Cui, Y., Wang, S., Jiang, L., Cao, J., & Zhu, S., et al. (2020). Calcium-binding protein S100P promotes tumor progression but enhances chemosensitivity in breast cancer. Frontiers in oncology, 10, 566302.
Goh, K. W., Stephen, A., Wu, Y. S., Sim, M. S., Batumalaie, K., & Gopinath, S. C., et al. (2023). Molecular targets of aptamers in gastrointestinal cancers: Cancer detection, therapeutic applications, and associated mechanisms. Journal of Cancer, 14, 2491.
Sun, W., Luo, L., Fang, D., Tang, T., Ni, W., & Dai, B., et al. (2020). A novel DNA aptamer targeting S100P induces antitumor effects in colorectal cancer cells. Nucleic acid therapeutics, 30, 402–413.
Möller, A., & Lobb, R. J. (2020). The evolving translational potential of small extracellular vesicles in cancer. Nature Reviews Cancer, 20, 697–709.
YoungHyeon, K., Cha, B. S., Park, K. S. (2023). Discovery of a Unique DNA Aptamer Directed at Small Extracellular Vesicles Originating from Colorectal Cancer Cells, Envisioned for Diagnostic and Therapeutic Implementations. 326.
Cha, B. S., Jang, Y. J., Lee, E. S., Kim, D. Y., Woo, J. S., & Son, J., et al. (2023). Development of a Novel DNA Aptamer Targeting Colorectal Cancer Cell‐Derived Small Extracellular Vesicles as a Potential Diagnostic and Therapeutic Agent. Advanced Healthcare Materials, 12, 2300854.
Song, L., Hao, Y., Wang, C., Han, Y., Zhu, Y., & Feng, L., et al. (2022). Liposomal oxaliplatin prodrugs loaded with metformin potentiate immunotherapy for colorectal cancer. Journal of Controlled Release, 350, 922–932.
Dearmond, S. J., Qiu, Y., Sanchez, H., Spilman, P. R., Ninchak-Casey, A., & Alonso, D., et al. (1999). PrPc glycoform heterogeneity as a function of brain region: implications for selective targeting of neurons by prion strains. Journal of neuropathology and experimental neurology, 58, 1000–1009.
Go, G., Lee, C.-S., Yoon, Y. M., Lim, J. H., Kim, T. H., & Lee, S. H. (2021). Prpc aptamer conjugated–gold nanoparticles for targeted delivery of doxorubicin to colorectal cancer cells. International Journal of Molecular Sciences, 22, 1976.
Lohlamoh, W., Soontornworajit, B., & Rotkrua, P. (2021). Anti-proliferative effect of doxorubicin-loaded AS1411 aptamer on colorectal cancer cell. Asian Pacific Journal of Cancer Prevention: APJCP, 22, 2209.
Zhu, H., Li, T., Du, Y., & Li, M. (2018). Pancreatic cancer: challenges and opportunities. BMC medicine, 16, 1–3.
Pereira, N. P., & Corrêa, J. R. (2018). Pancreatic cancer: Treatment approaches and trends. J Cancer Metastasis Treat, 4, 10.20517.
Jimeno, A., Tan, A. C., Coffa, J., Rajeshkumar, N., Kulesza, P., & Rubio-Viqueira, B., et al. (2008). Coordinated epidermal growth factor receptor pathway gene overexpression predicts epidermal growth factor receptor inhibitor sensitivity in pancreatic cancer. Cancer research, 68, 2841–2849.
Mahajan, U. M., Li, Q., Alnatsha, A., Maas, J., Orth, M., & Maier, S. H., et al. (2021). Tumor-specific delivery of 5-fluorouracil–incorporated epidermal growth factor receptor–targeted aptamers as an efficient treatment in pancreatic ductal adenocarcinoma models. Gastroenterology, 161, 996–1010.e1.
Heinemann, V. (2000). Gemcitabine: progress in the treatment of pancreatic cancer. Oncology, 60, 8–18.
Hong, S. S., Lee, S., Lee, S. H., Kim, S., Kim, D., & Park, H., et al. (2022). Anticancer effect of locally applicable aptamer‐conjugated gemcitabine‐loaded atelocollagen patch in pancreatic cancer patient–derived xenograft models. Cancer Science, 113, 1752–1762.
Choi, S. I., Lee, Y.-S., Lee, Y. M., Kim, H. J., Kim, W. J., & Jung, S., et al. (2023). Complexation of drug and hapten-conjugated aptamer with universal hapten antibody for pancreatic cancer treatment. Journal of Controlled Release, 360, 940–952.
Bengoechea-Alonso, M. T., & Ericsson, J. (2010). The ubiquitin ligase Fbxw7 controls adipocyte differentiation by targeting C/EBPα for degradation. Proceedings of the National Academy of Sciences, 107, 11817–11822.
Zheng, L., Wang, L., Gan, J., & Zhang, H. (2014). RNA activation: promise as a new weapon against cancer. Cancer Letters, 355, 18–24.
Halama, N., Prüfer, U., Frömming, A., Beyer, D., Eulberg, D., & Jungnelius, J. U., et al. (2019). Phase I/II study with CXCL12 inhibitor NOX-A12 and pembrolizumab in patients with microsatellite-stable, metastatic colorectal or pancreatic cancer. Annals of Oncology, 30, v231.
Yoon, S., Huang, K.-W., Reebye, V., Mintz, P., Tien, Y.-W., & Lai, H.-S., et al. (2016). Targeted delivery of C/EBPα-saRNA by pancreatic ductal adenocarcinoma-specific RNA aptamers inhibits tumor growth in vivo. Molecular Therapy, 24, 1106–1116.
Ozakyol, A. (2017). Global epidemiology of hepatocellular carcinoma (HCC epidemiology). Journal of gastrointestinal cancer, 48, 238–240.
Iliaki, S., Beyaert, R., & Afonina, I. S. (2021). Polo-like kinase 1 (PLK1) signaling in cancer and beyond. Biochemical pharmacology, 193, 114747.
Yu, X.-X., Ge, K.-L., Liu, N., Zhang, J.-Y., Xue, M.-L., Ge, Y.-L. (2020). Selection and characterization of a novel DNA aptamer, Apt-07S specific to hepatocellular carcinoma cells. Drug Design, Development Therapy, 14, 1535–1545.
Ganesan, S., & Mehnert, J. (2020). Biomarkers for response to immune checkpoint blockade. Annual Review of Cancer Biology, 4, 331–351.
Ott, P. A., Hodi, F. S., & Robert, C. (2013). CTLA-4 and PD-1/PD-L1 blockade: new immunotherapeutic modalities with durable clinical benefit in melanoma patients. Clinical cancer research, 19, 5300–5309.
Du, Y., Zhang, D., Wang, Y., Wu, M., Zhang, C., & Zheng, Y., et al. (2021). A highly stable multifunctional aptamer for enhancing antitumor immunity against hepatocellular carcinoma by blocking dual immune checkpoints. Biomaterials Science, 9, 4159–4168.
Chakraborty, S., Dlie, Z. Y., Chakraborty, S., Roy, S., Mukherjee, B., & Besra, S. E., et al. (2020). Aptamer-functionalized drug nanocarrier improves hepatocellular carcinoma toward normal by targeting neoplastic hepatocytes. Molecular Therapy-Nucleic Acids, 20, 34–49.
Zhang, L., Zhou, L., Zhang, H., Zhang, Y., Li, L., & **e, T., et al. (2021). Development of a DNA aptamer against multidrug-resistant hepatocellular carcinoma for in vivo imaging. ACS Applied Materials & Interfaces, 13, 54656–54664.
Huang, B. T., Lai, W. Y., Chang, Y. C., Wang, J. W., Yeh, S. D., & Lin, E. P., et al. (2017). A CTLA-4 Antagonizing DNA Aptamer with Antitumor Effect. Molecular therapy Nucleic acids, 8, 520–528.
Herrmann, A., Priceman, S. J., Swiderski, P., Kujawski, M., **n, H., & Cherryholmes, G. A., et al. (2014). CTLA4 aptamer delivers STAT3 siRNA to tumor-associated and malignant T cells. The Journal of clinical investigation, 124, 2977–2987.
Santulli-Marotto, S., Nair, S. K., Rusconi, C., Sullenger, B., & Gilboa, E. (2003). Multivalent RNA aptamers that inhibit CTLA-4 and enhance tumor immunity. Cancer research, 63, 7483–7489.
Lai, W. Y., Huang, B. T., Wang, J. W., Lin, P. Y., & Yang, P. C. (2016). A Novel PD-L1-targeting Antagonistic DNA Aptamer With Antitumor Effects. Molecular therapy Nucleic acids, 5, e397.
Schrand, B., Berezhnoy, A., Brenneman, R., Williams, A., Levay, A., & Kong, L. Y., et al. (2014). Targeting 4-1BB costimulation to the tumor stroma with bispecific aptamer conjugates enhances the therapeutic index of tumor immunotherapy. Cancer immunology research, 2, 867–877.
An, Y., Li, X., Yao, F., Duan, J. & Yang, X. D. (2022). Novel Complex of PD-L1 Aptamer and Albumin Enhances Antitumor Efficacy In Vivo. Molecules (Basel, Switzerland), 27, 1482.
McNamara, J. O., Kolonias, D., Pastor, F., Mittler, R. S., Chen, L., & Giangrande, P. H., et al. (2008). Multivalent 4-1BB binding aptamers costimulate CD8+ T cells and inhibit tumor growth in mice. The Journal of clinical investigation, 118, 376–386.
Soldevilla, M. M., Villanueva, H., Bendandi, M., Inoges, S., López-Díaz de Cerio, A., & Pastor, F. (2015). 2-fluoro-RNA oligonucleotide CD40 targeted aptamers for the control of B lymphoma and bone-marrow aplasia. Biomaterials, 67, 274–285.
Takahashi, M., Hashimoto, Y., & Nakamura, Y. (2022). Anti-TGF-β1 aptamer enhances therapeutic effect of tyrosine kinase inhibitor, gefitinib, on non-small cell lung cancer in xenograft model. Molecular therapy Nucleic acids, 29, 969–978.
Author information
Authors and Affiliations
Contributions
"E.F, S.V and A.H. wrote the main manuscript text and F.M, M.K and A.Z prepared figures 1 and Tables. M.D, M.S and A.H reviewed the final version. H.U. supervision. All authors reviewed the manuscript."
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher’s note Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Uinarni, H., Oghenemaro, E.F., Menon, S.V. et al. Breaking Barriers: Nucleic Acid Aptamers in Gastrointestinal (GI) Cancers Therapy. Cell Biochem Biophys (2024). https://doi.org/10.1007/s12013-024-01367-w
Accepted:
Published:
DOI: https://doi.org/10.1007/s12013-024-01367-w