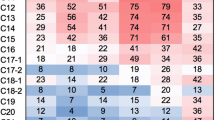
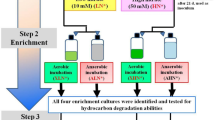
Abstract
Oil sands tailings, a slurry of alkaline water, silt, clay, unrecovered bitumen, and residual hydrocarbons generated during bitumen extraction, are contained in ponds. Indigenous microbes metabolize hydrocarbons and emit greenhouse gases from the tailings. Metabolism of hydrocarbons in tailings ponds of two operators, namely, Canadian Natural Upgrading Limited (CNUL) and Canadian Natural Resources Limited (CNRL), has not been comprehensively investigated. Previous reports have revealed sequential and preferential hydrocarbon degradation of alkanes in primary cultures established from CNUL and CNRL tailings amended separately with mixtures of hydrocarbons (n-alkanes, iso-alkanes, paraffinic solvent, or naphtha). In this study, activation pathway of hydrocarbon biodegradation in these primary cultures was investigated. The functional gene analysis revealed that fumarate addition was potentially the primary activation pathway of alkanes in all cultures. However, the metabolite analysis only detected transient succinylated 2-methylpentane and 2-methylbutane metabolites during initial methanogenic biodegradation of iso-alkanes and paraffinic solvent in all CNUL and CNRL cultures amended with iso-alkanes and paraffinic solvent. Under sulfidogenic conditions (prepared only with CNUL tailings amended with iso-alkanes), succinylated 2-methylpentane persisted throughout incubation period of ~ 1100 days, implying dead-end nature of the metabolite. Though no metabolite was detected in n-alkanes- and naphtha-amended cultures during incubation, assA/masD genes related to Peptococcaceae were amplified in all CNUL and CNRL primary cultures. The findings of this present study suggest that microbial communities in different tailings ponds can biodegrade hydrocarbons through fumarate addition as activation pathway under methanogenic and sulfidogenic conditions.


Similar content being viewed by others
Data Availability
All data generated or analyzed during this study are included in this published article and its supplementary information files.
References
Blanksby, S. J., & Ellison, G. B. (2003). Bond dissociation energies of organic molecules. Accounts of chemical research, 36(4), 255–263. https://doi.org/10.1021/ar020230d
Boll, M., & Heider, J. (2010). Anaerobic degradation of hydrocarbons: Mechanisms of C-H-bond activation in the absence of oxygen. In Handbook of Hydrocarbon and Lipid Microbiology (Vol. 1, pp. 1011–1024). Springer. https://doi.org/10.1007/978-3-540-77587-4
Callaghan, A. V., Wawrik, B., Ní Chadhain, S. M., Young, L. Y., & Zylstra, G. J. (2008). Anaerobic alkane-degrading strain AK-01 contains two alkylsuccinate synthase genes. Biochemical and biophysical research communications, 366(1), 142–148. https://doi.org/10.1016/j.bbrc.2007.11.094
Zedelius, J., Rabus, R., Grundmann, O., Werner, I., Brodkorb, D., Schreiber, F., … Widdel, F. (2011). Alkane degradation under anoxic conditions by a nitrate-reducing bacterium with possible involvement of the electron acceptor in substrate activation. Environmental Microbiology Reports, 3(1), 125–135. https://doi.org/10.1111/j.1758-2229.2010.00198.x
Khelifi, N., Amin Ali, O., Roche, P., Grossi, V., Brochier-Armanet, C., Valette, O., … Hirschler-Réa, A. (2014). Anaerobic oxidation of long-chain n-alkanes by the hyperthermophilic sulfate-reducing archaeon, Archaeoglobus fulgidus. The ISME Journal, 8(11), 2153–2166. https://doi.org/10.1038/ismej.2014.58
Zhou, L., Li, K.-P., Mbadinga, S. M., Yang, S.-Z., Gu, J.-D., & Mu, B.-Z. (2012). Analyses of n-alkanes degrading community dynamics of a high-temperature methanogenic consortium enriched from production water of a petroleum reservoir by a combination of molecular techniques. Ecotoxicology, 21(6), 1680–1691. https://doi.org/10.1007/s10646-012-0949-5
Bian, X.-Y., Maurice Mbadinga, S., Liu, Y.-F., Yang, S.-Z., Liu, J.-F., Ye, R.-Q., … Mu, B.-Z. (2015). Insights into the anaerobic biodegradation pathway of n-alkanes in oil reservoirs by detection of signature metabolites. Scientific Reports, 5, 9801. https://doi.org/10.1038/srep09801
Abu Laban, N., Dao, A., Semple, K., & Foght, J. M. (2015). Biodegradation of C7 and C8 iso-alkanes under methanogenic conditions. Environmental Microbiology, 17(12), 4898–4915. https://doi.org/10.1111/1462-2920.12643
Tan, B., Semple, K., & Foght, J. M. (2015). Anaerobic alkane biodegradation by cultures enriched from oil sands tailings ponds involves multiple species capable of fumarate addition. FEMS Microbiology Ecology, 91(5), iv042. https://doi.org/10.1093/femsec/fiv042
Beller, H. R., & Spormann, A. M. (1998). Analysis of the novel benzylsuccinate synthase reaction for anaerobic toluene activation based on structural studies of the product. Journal of Bacteriology, 180(20), 5454–5457. Retrieved from http://www.pubmedcentral.nih.gov/articlerender.fcgi?artid=107597&tool=pmcentrez&rendertype=abstract
Bozinovski, D., Herrmann, S., Richnow, H.-H., von Bergen, M., Seifert, J., & Vogt, C. (2012). Functional analysis of an anaerobic m-xylene-degrading enrichment culture using protein-based stable isotope probing. FEMS Microbiology Ecology, 81(1), 134–144. https://doi.org/10.1111/j.1574-6941.2012.01334.x
Fowler, S. J., Dong, X., Sensen, C. W., Suflita, J. M., & Gieg, L. M. (2012). Methanogenic toluene metabolism: community structure and intermediates. Environmental microbiology, 14(3), 754–764. https://doi.org/10.1111/j.1462-2920.2011.02631.x
Gieg, L. M., & Suflita, J. M. (2002). Detection of anaerobic metabolites of saturated and aromatic hydrocarbons in petroleum-contaminated aquifers. Environmental Science & Technology, 36(17), 3755–3762. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/12322748
Selesi, D., Jehmlich, N., Von Bergen, M., Schmidt, F., Rattei, T., Tischler, P., … Meckenstock, R. U. (2010). Combined genomic and proteomic approaches identify gene clusters involved in anaerobic 2-methylnaphthalene degradation in the sulfate-reducing enrichment culture N47. Journal of Bacteriology, 192(1), 295–306. https://doi.org/10.1128/JB.00874-09
Berdugo-Clavijo, C., Dong, X., Soh, J., Sensen, C. W., & Gieg, L. M. (2012). Methanogenic biodegradation of two-ringed polycyclic aromatic hydrocarbons. FEMS Microbiology Ecology, 81(1), 124–133. https://doi.org/10.1111/j.1574-6941.2012.01328.x
Annweiler, E., Materna, A., Safinowski, M., Kappler, A., Richnow, H. H., Michaelis, W., & Meckenstock, R. U. (2000). Anaerobic degradation of 2-methylnaphthalene by a sulfate-reducing enrichment culture. Applied and Environmental Microbiology, 66(12), 5329–5333. https://doi.org/10.1128/AEM.66.12.5329-5333.2000.Updated
Callaghan, A. V. (2013). Enzymes involved in the anaerobic oxidation of n-alkanes: From methane to long-chain paraffins. Frontiers in Microbiology, 4, 89. https://doi.org/10.3389/fmicb.2013.00089
Siddique, T., Penner, T., Klassen, J., Nesbø, C., & Foght, J. M. (2012). Microbial communities involved in methane production from hydrocarbons in oil sands tailings. Environmental Science & Technology, 46(17), 9802–9810. https://doi.org/10.1021/es302202c
Siddique, T., Fedorak, P. M., MacKinnon, M. D., & Foght, J. M. (2007). Metabolism of BTEX and naphtha compounds to methane in oil sands tailings. Environmental Science & Technology, 41(7), 2350–2356. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/17438786
Siddique, T., Fedorak, P. M., & Foght, J. M. (2006). Biodegradation of short-chain n-alkanes in oil sands tailings under methanogenic conditions. Environmental Science & Technology, 40(17), 5459–5464. Retrieved from http://www.ncbi.nlm.nih.gov/pubmed/16999125
Siddique, T., Semple, K., Li, C., & Foght, J. M. (2020). Methanogenic biodegradation of iso-alkanes and cycloalkanes during long-term incubation with oil sands tailings. Environmental Pollution, 258. https://doi.org/10.1016/j.envpol.2019.113768
Kong, J. D., Wang, H., Siddique, T., Foght, J., Semple, K., Burkus, Z., & Lewis, M. A. (2019). Second-generation stoichiometric mathematical model to predict methane emissions from oil sands tailings. Science of the Total Environment, 694, 133645. https://doi.org/10.1016/j.scitotenv.2019.133645
Mohamad Shahimin, M. F., & Siddique, T. (2017). Methanogenic biodegradation of paraffinic solvent hydrocarbons in two different oil sands tailings. Science of the Total Environment, 583, 115–122. https://doi.org/10.1016/j.envpol.2016.12.002
Mohamad Shahimin, M. F., & Siddique, T. (2017). Sequential biodegradation of complex naphtha hydrocarbons under methanogenic conditions in two different oil sands tailings. Environmental Pollution, 221, 398–406. https://doi.org/10.1016/j.envpol.2016.12.002
Mohamad Shahimin, M. F., Foght, J. M., & Siddique, T. (2016). Preferential methanogenic biodegradation of short-chain n-alkanes by microbial communities from two different oil sands tailings ponds. Science of Total Environment, 553, 250–257. https://doi.org/10.1016/j.scitotenv.2016.02.061
Mohamad Shahimin, M. F., Foght, J. M., & Siddique, T. (2021). Methanogenic biodegradation of iso-alkanes by indigenous microbes from two different oil sands tailings ponds. Microorganisms, 9(8), 1569. https://doi.org/10.3390/microorganisms9081569
Mohamad Shahimin, M. F., & Siddique, T. (2023). Biodegradation of 2-methylpentane in fluid fine tailings amended with a mixture of iso -alkanes under sulfate-reducing conditions. Canadian Journal of Microbiology. https://doi.org/10.1139/cjm-2023-0022
Mohamad Shahimin, M. F. (2016). Anaerobic biodegradation of hydrocarbons in different oil sands tailings ponds: Key microbial players and main activation pathway of hydrocarbon biodegradation. University of Alberta.
So, C. M., Phelps, C. D., & Young, L. Y. (2003). Anaerobic transformation of alkanes to fatty acids by a sulfate-reducing anaerobic transformation of alkanes to fatty acids by a sulfate-reducing bacterium, strain Hxd3. Applied and Environmental Microbiology, 69(7), 3892–3900. https://doi.org/10.1128/AEM.69.7.3892
Kropp, K. G., Davidova, I. A., & Suflita, J. M. (2000). Anaerobic oxidation of n-dodecane by an addition reaction in a sulfate-reducing bacterial enrichment culture. Applied and Environmental Microbiology, 66(12), 5393–5398. https://doi.org/10.1128/AEM.66.12.5393-5398.2000
Callaghan, A. A. V, Davidova, I. A., Savage-Ashlock, K., Parisi, V. A., Gieg, L. M., Suflita, J. M., … Wawrik, B. (2010). Diversity of benzyl- and alkylsuccinate synthase genes in hydrocarbon-impacted environments and enrichment cultures. Environmental Science & Technology, 44(19), 7287–7294. https://doi.org/10.1021/es1002023
Winderl, C., Schaefer, S., & Lueders, T. (2007). Detection of anaerobic toluene and hydrocarbon degraders in contaminated aquifers using benzylsuccinate synthase (bssA) genes as a functional marker. Environmental Microbiology, 9(4), 1035–1046. https://doi.org/10.1111/j.1462-2920.2006.01230.x
Edgar, R. C. (2004). MUSCLE: a multiple sequence alignment method with reduced time and space complexity. BMC Bioinformatics, 5, 113. https://doi.org/10.1186/1471-2105-5-113
Guindon, S., Gascuel, O., Dufayard, J.-F., Lefort, V., Anisimova, M., & Hordijk, W. (2010). New algorithms and methods to estimate maximim-likelihood phylogenies: assessing the performance of PhyML 3.0. Systematic Biology, 59(3), 307–321. https://doi.org/10.1093/sysbio/syq010
Abu Laban, N., Tan, B., Dao, A., & Foght, J. M. (2015). Draft genome sequence of uncultivated toluene-degrading Desulfobulbaceae bacterium Tol-SR, obtained by stable isotope probing using [13 C 6 ] toluene. Genome Announcements, 3(1), e01423-e1514. https://doi.org/10.1128/genomeA.01423-14
Toth, C. R. A., & Gieg, L. M. (2018). Time course-dependent methanogenic crude oil biodegradation: dynamics of fumarate addition metabolites, biodegradative genes, and microbial community composition. Frontiers in Microbiology, 8(JAN), 1–16. https://doi.org/10.3389/fmicb.2017.02610
Liu, J. F., Lu, Y. W., Zhou, L., Li, W., Hou, Z. W., Yang, S. Z., … Mu, B. Z. (2020). Simultaneous detection of transcribed functional assA gene and the corresponding metabolites of linear alkanes (C4, C5, and C7) in production water of a low-temperature oil reservoir. Science of the Total Environment, 746, 141290. https://doi.org/10.1016/j.scitotenv.2020.141290
Chen, J., Zhou, L., Liu, Y. F., Hou, Z. W., Li, W., Mbadinga, S. M., & Mu, B. Z. (2020). Synthesis and mass spectra of rearrangement bio-signature metabolites of anaerobic alkane degradation via fumarate addition. Analytical Biochemistry, 600(March), 113746. https://doi.org/10.1016/j.ab.2020.113746
Tan, B., Charchuk, R., Li, C., Abu Laban, N., & Foght, J. M. (2014). Draft genome sequence of uncultivated firmicutes (Peptococcaceae SCADC) single cells sorted from methanogenic alkane-degrading cultures. Genome Announcements, 2(5), e00909-e914. https://doi.org/10.1128/genomeA.00909-14
Foght, J. M. (2008). Anaerobic biodegradation of aromatic hydrocarbons: pathways and prospects. Journal of molecular microbiology and biotechnology, 15(2–3), 93–120. https://doi.org/10.1159/000121324
Musat, F. (2015). The anaerobic degradation of gaseous, nonmethane alkanes - from in situ processes to microorganisms. Computational and Structural Biotechnology Journal, 13, 222–228. https://doi.org/10.1016/j.csbj.2015.03.002
Ji, J. H., Zhou, L., Mbadinga, S. M., Irfan, M., Liu, Y. F., Pan, P., … Mu, B. Z. (2020). Methanogenic biodegradation of C9 to C12 n-alkanes initiated by Smithella via fumarate addition mechanism. AMB Express, 10(1). https://doi.org/10.1186/s13568-020-0956-5
Ji, J. H., Liu, Y. F., Zhou, L., Mbadinga, S. M., Pan, P., Chen, J., … Mu, B. Z. (2019). Methanogenic degradation of long n-alkanes requires fumarate-dependent activation. Applied and Environmental Microbiology, 85(16), e00985–19. https://doi.org/10.1128/AEM.00985-19
Oberding, L. K., & Gieg, L. M. (2018). Methanogenic paraffin biodegradation: alkylsuccinate synthase gene quantification and dicarboxylic acid production. Applied and Environmental Microbiology, 84(1), e01773-e1817. https://doi.org/10.1128/AEM.01773-17
Aitken, C. M., Jones, D. M., Maguire, M. J., Gray, N. D., Sherry, A., Bowler, B. F. J., … Head, I. M. (2013). Evidence that crude oil alkane activation proceeds by different mechanisms under sulfate-reducing and methanogenic conditions. Geochimica et Cosmochimica Acta, 109, 162–174. https://doi.org/10.1016/j.gca.2013.01.031
Jarling, R., Sadeghi, M., Drozdowska, M., Lahme, S., Buckel, W., Rabus, R., … Wilkes, H. (2012). Stereochemical investigations reveal the mechanism of the bacterial activation of n-alkanes without oxygen. Angewandte Chemie - International Edition, 51(6), 1334–1338. https://doi.org/10.1002/anie.201106055
Chen, J., Liu, Y., Zhou, L., Mbadinga, S. M., Yang, T., Zhou, J., & Liu, J. (2019). Methanogenic degradation of branched alkanes in enrichment cultures of production water from a high-temperature petroleum reservoir. Applied Microbiology and Biotechnology, 1–4. https://doi.org/10.1007/s00253-018-09574-1
Siddique, T., Mohamad Shahimin, M. F. M. F., Zamir, S., Semple, K., Li, C., & Foght, J. M. J. M. (2015). Long-term incubation reveals methanogenic biodegradation of C5 and C6 iso-alkanes in oil sands tailings. Environmental Science & Technology, 49(24), 14732–14739. https://doi.org/10.1021/acs.est.5b04370
An, D., Brown, D., Chatterjee, I., Dong, X., Ramos-Padron, E., Wilson, S., … Voordouw, G. (2013). Microbial community and potential functional gene diversity involved in anaerobic hydrocarbon degradation and methanogenesis in an oil sands tailings pond. Genome, 56(10), 612–618. https://doi.org/10.1139/gen-2013-0083
Acknowledgements
We thank Canadian Natural Resources Ltd. and Shell CNUL Sands Inc. for providing oil sands tailings samples.
Funding
We gratefully acknowledge funding from the Helmholtz-Alberta Initiative (TS), NSERC Discovery Grant (TS), and Canada Foundation for Innovation (TS). We also acknowledge a Ph.D. scholarship award (King’s Scholarship) and fellowship to MFMS from the Public Service Department of Malaysia and Universiti Malaysia Perlis, respectively.
Author information
Authors and Affiliations
Contributions
Conceptualization: TS and MFMS, data curation: MFMS, formal analysis: MFMS, funding acquisition: TS, investigation: MFMS, methodology: MFMS, project administration: MFMS, resources: TS, supervision: TS, validation: TS, writing original draft: MFMS, and review and editing: MFMS and TS.
Corresponding author
Ethics declarations
Ethics Approval
Not applicable.
Consent to Participate
Not applicable.
Consent for Publication
Not applicable.
Competing Interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Mohamad Shahimin, M.F., Siddique, T. Uncovering Anaerobic Hydrocarbon Biodegradation Pathways in Oil Sands Tailings from Two Different Tailings Ponds via Metabolite and Functional Gene Analyses. Appl Biochem Biotechnol (2024). https://doi.org/10.1007/s12010-024-04855-0
Accepted:
Published:
DOI: https://doi.org/10.1007/s12010-024-04855-0