Abstract
The continued spread of SARS-CoV-2 has presented unprecedented obstacles to the worldwide public health system. Especially, individuals with chronic obstructive pulmonary disease (COPD) are at a heightened risk of contracting SARS-CoV-2 infection due to their pre-existing respiratory symptoms that are not well-managed. However, the viral mechanism of affecting the expression of host genes, COPD progression, and prognosis is not clear yet.
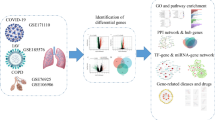
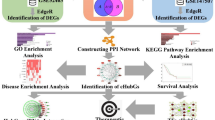
This study integrated the differential expression information of COPD patients and then calculated the correlation between mRNAs and miRNAs to construct a COPD-specific ceRNA network. The DEGs of individuals with SARS-CoV-2 infection and anticipated miRNAs and their targets were analyzed in 9 SARS-CoV-2 sequences from different geographic locations. Furthermore, combining the experimentally validated miRNAs and genes, the regulatory miRNA-mRNA relationships were identified. All the regulatory relationships were integrated into the COPD-specific network and the network modules were explored to get insight into the functional mechanism of SARS-CoV-2 infection in COPD patients.
A higher proportion of DEGs compete with the same miRNA suggesting a higher expression of genes in the COPD-specific ceRNA network. Hsa-miR-21-3p is the largest connected point in the network, but the proportion of genes upregulated by hsa-miR-21-3p is low (P = 0.1406). This indicates that the regulatory relationship of competitive inhibition has little effect on has-miR-21, and the high expression pattern is a poor prognostic factor in COPD. Hsa-miR-15a-5p is the most significant miRNA with the highest proportion of DEGs. And ANXA2P3 is the only gene in the COPD ceRNA network that interferes with hsa-miR-15a-5p. In addition, we found that has-miR-1184- and has-miR-99-cored modules were significant, and genes ZDHHC18, PCGF3, and KIAA0319L interacting with them were all associated with COPD prognosis, and high expression of these genes could lead to poor prognosis in COPD.
The key regulators such as miR-21, miR-15a, ANXA2P3, ZDHHC18, PCGF3, and KIAA0319L can be used as prognostic biomarkers for early intervention in COPD with SARS-CoV-2 infection.






Similar content being viewed by others
Data Availability
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Abbreviations
- ceRNA:
-
Competing endogenous ribonucleic acid
- COPD:
-
Chronic obstructive pulmonary disease
- SARS-CoV-2:
-
Severe acquired respiratory syndrome coronavirus 2
- miRNA:
-
MicroRNA
- DEGs:
-
Differentially expressed genes
References
GBD 2017 Causes of Death Collaborators. Global, regional, and national age-sex-specific mortality for 282 causes of death in 195 countries and territories, 1980–2017: A systematic analysis for the Global Burden of Disease Study 2017 [published correction appears in Lancet 2019 Jun 22;393(10190):e44] [published correction appears in Lancet. 2018 Nov 17;392(10160):2170]. Lancet. 2018;392(10159):1736–1788
Labaki, W. W., & Rosenberg, S. R. (2020). Chronic obstructive pulmonary disease. Annals of Internal Medicine, 173(3), ITC17–ITC32. https://doi.org/10.7326/AITC202008040
Cho, M. H., McDonald, M. L., Zhou, X., et al. (2014). Risk loci for chronic obstructive pulmonary disease: A genome-wide association study and meta-analysis. The Lancet Respiratory Medicine, 2(3), 214–225.
Hobbs, B. D., de Jong, K., Lamontagne, M., et al. (2017). Genetic loci associated with chronic obstructive pulmonary disease overlap with loci for lung function and pulmonary fibrosis. Nature Genetics, 49(3), 426–432.
Sakornsakolpat, P., Prokopenko, D., Lamontagne, M., et al. (2019). Genetic landscape of chronic obstructive pulmonary disease identifies heterogeneous cell-type and phenotype associations. Nature Genetics, 51(3), 494–505.
Manichaikul, A., Hoffman, E. A., Smolonska, J., et al. (2014). Genome-wide study of percent emphysema on computed tomography in the general population. The Multi-Ethnic Study of Atherosclerosis Lung/SNP Health Association Resource Study. American Journal of Respiratory and Critical Care Medicine, 189(4), 408–418.
Boueiz, A., Lutz, S. M., Cho, M. H., et al. (2017). Genome-wide association study of the genetic determinants of emphysema distribution. American Journal of Respiratory and Critical Care Medicine, 195(6), 757–771.
Wittstatt, J., Weider, M., Wegner, M., & Reiprich, S. (2020). MicroRNA miR-204 regulates proliferation and differentiation of oligodendroglia in culture. Glia, 68(10), 2015–2027.
Wu, B., Liu, G., **, Y., et al. (2020). miR-15b-5p promotes growth and metastasis in breast cancer by targeting HPSE2. Frontiers in Oncology, 10, 108.
Devadoss, D., Long, C., Langley, R. J., et al. (2019). Long noncoding transcriptome in chronic obstructive pulmonary disease. American Journal of Respiratory Cell and Molecular Biology, 61(6), 678–688.
Gu, W., Yuan, Y., Wang, L., et al. (2019). Long non-coding RNA TUG1 promotes airway remodelling by suppressing the miR-145-5p/DUSP6 axis in cigarette smoke-induced COPD. Journal of Cellular and Molecular Medicine, 23(11), 7200–7209.
Khailany, R. A., Safdar, M., & Ozaslan, M. (2020). Genomic characterization of a novel SARS-CoV-2. Gene Rep., 19, 100682.
Zhou, P., Yang, X. L., Wang, X. G., et al. (2020). A pneumonia outbreak associated with a new coronavirus of probable bat origin [published correction appears in Nature. 2020 Dec; 588(7836):E6]. Nature, 579(7798), 270–273.
Chen, B., Tian, E. K., He, B., et al. (2020). Overview of lethal human coronaviruses. Signal Transduction and Targeted Therapy, 5(1), 89.
Evans, S. E., Tseng, C. K., Scott, B. L., Höök, A. M., & Dickey, B. F. (2020). Inducible epithelial resistance against coronavirus pneumonia in mice. American Journal of Respiratory Cell and Molecular Biology, 63(4), 540–541.
Kincaid, R. P., & Sullivan, C. S. (2012). Virus-encoded microRNAs: An overview and a look to the future. PLoS Pathogens, 8(12), e1003018.
Rastogi, M., Pandey, N., Shukla, A., & Singh, S. K. (2020). SARS coronavirus 2: From genome to infectome. Respiratory Research, 21(1), 318.
Leung, J. M., Yang, C. X., Tam, A., et al. (2020). ACE-2 expression in the small airway epithelia of smokers and COPD patients: Implications for COVID-19. European Respiratory Journal, 55(5), 2000688.
Barrett, T., Wilhite, S. E., Ledoux, P., et al. (2013). NCBI GEO: Archive for functional genomics data sets–update. Nucleic Acids Research, 41(Database issue), 991–995.
Kozomara, A., Birgaoanu, M., & Griffiths-Jones, S. (2019). miRBase: From microRNA sequences to function. Nucleic Acids Research, 47(D1), D155–D162.
Stelzer, G., Rosen, N., Plaschkes, I., et al. (2016). The GeneCards suite: From gene data mining to disease genome sequence analyses. Current Protocols in Bioinformatics, 54, 1.30.1-1.30.33.
Hsu, S. D., Tseng, Y. T., Shrestha, S., et al. (2014). miRTarBase update 2014: An information resource for experimentally validated miRNA-target interactions. Nucleic Acids Research, 42(1), 78–85.
Larkin, M. A., Blackshields, G., Brown, N. P., et al. (2007). Clustal W and Clustal X version 2.0. Bioinformatics, 23(21), 2947–2948.
Kumar, S., Stecher, G., Li, M., Knyaz, C., & Tamura, K. (2018). MEGA X: Molecular evolutionary genetics analysis across computing platforms. Molecular Biology and Evolution, 35(6), 1547–1549.
Lukasik, A., Wójcikowski, M., & Zielenkiewicz, P. (2016). Tools4miRs - One place to gather all the tools for miRNA analysis. Bioinformatics, 32(17), 2722–2724.
Fabian, M. R., Sonenberg, N., & Filipowicz, W. (2010). Regulation of mRNA translation and stability by microRNAs. Annual Review of Biochemistry, 79, 351–379.
Thomson, D. W., & Dinger, M. E. (2016). Endogenous microRNA sponges: Evidence and controversy. Nature Reviews Genetics, 17(5), 272–283.
Gao, Z., Liu, H., Shi, Y., Yin, L., Zhu, Y., & Liu, R. (2019). Identification of cancer stem cell molecular markers and effects of hsa-miR-21-3p on stemness in esophageal squamous cell carcinoma. Cancers (Basel), 11(4), 518.
He, S., Sun, S., Lu, J., et al. (2021). The effects of the miR-21/SMAD7/TGF-β pathway on Th17 cell differentiation in COPD. Science and Reports, 11(1), 6338.
Kim, R. Y., Sunkara, K. P., Bracke, K. R., et al. (2021). A microRNA-21-mediated SATB1/S100A9/NF-κB axis promotes chronic obstructive pulmonary disease pathogenesis. Science Translational Medicine, 13(621), eaav7223.
Lu, J., **e, L., & Sun, S. (2021). The inhibitor miR-21 regulates macrophage polarization in an experimental model of chronic obstructive pulmonary disease. Tobacco Induced Diseases, 19, 69.
Xu, H., Ling, M., Xue, J., et al. (2018). Exosomal microRNA-21 derived from bronchial epithelial cells is involved in aberrant epithelium-fibroblast cross-talk in COPD induced by cigarette smoking. Theranostics, 8(19), 5419–5433.
Drake, J., McMichael, G. O., Vornholt, E. S., et al. (2020). Assessing the role of long noncoding RNA in nucleus accumbens in subjects with alcohol dependence. Alcoholism, Clinical and Experimental Research, 44(12), 2468–2480.
Qian, Y., Mao, Z. D., Shi, Y. J., Liu, Z. G., Cao, Q., & Zhang, Q. (2018). Comprehensive analysis of miRNA-mRNA-lncRNA networks in non-smoking and smoking patients with chronic obstructive pulmonary disease. Cellular Physiology and Biochemistry, 50(3), 1140–1153.
Ju, C. R., **a, X. Z., & Chen, R. C. (2007). Expressions of tumor necrosis factor-converting enzyme and ErbB3 in rats with chronic obstructive pulmonary disease. Chinese Med J-Peking, 120(17), 1505–1510.
Yu, H., Li, Q., Kolosov, V. P., Perelman, J. M., & Zhou, X. (2011). Regulation of cigarette smoke-induced mucin expression by neuregulin1β/ErbB3 signalling in human airway epithelial cells. Basic & Clinical Pharmacology & Toxicology, 109(1), 63–72.
Nouws, J., Wan, F., Finnemore, E., et al. (2021). MicroRNA miR-24-3p reduces DNA damage responses, apoptosis, and susceptibility to chronic obstructive pulmonary disease. JCI Insight., 6(2), e134218.
Hu, Y., Cheng, Y., Jiang, X., et al. (2021). PCGF3 promotes the proliferation and migration of non-small cell lung cancer cells via the PI3K/AKT signaling pathway. Experimental Cell Research, 400(2), 112496.
Chen, Q., Zhang, H., Zhang, J., et al. (2021). miR-210-3p promotes lung cancer development and progression by modulating USF1 and PCGF3. Oncotargets and Therapy, 14, 3687–3700.
Zhu, M., Ye, M., Wang, J., Ye, L., & **, M. (2020). Construction of potential miRNA-mRNA regulatory network in COPD plasma by bioinformatics analysis. International Journal of Chronic Obstructive Pulmonary Disease, 15, 2135–2145.
**e, T. A., Han, M. Y., Su, X. R., Li, H. H., Chen, J. C., & Guo, X. G. (2020). Identification of Hub genes associated with infection of three lung cell lines by SARS-CoV-2 with integrated bioinformatics analysis. Journal of Cellular and Molecular Medicine, 24(20), 12225–12230.
García-Expósito, L., Barroso-González, J., Puigdomènech, I., Machado, J. D., Blanco, J., & Valenzuela-Fernández, A. (2011). HIV-1 requires Arf6-mediated membrane dynamics to efficiently enter and infect T lymphocytes. Molecular Biology of the Cell, 22(8), 1148–1166.
Haqshenas, G., & Doerig, C. (2019). Targeting of host cell receptor tyrosine kinases by intracellular pathogens. Science Signaling, 12(599), eaau9894.
Soroceanu, L., Akhavan, A., & Cobbs, C. S. (2008). Platelet-derived growth factor-alpha receptor activation is required for human cytomegalovirus infection. Nature, 455(7211), 391–395.
Acknowledgements
We are thankful to Dr. Lindong Yuan, for critically editing the current manuscript.
Funding
This work was funded by the 2021 Annual Science and Technology Development Plan of Shandong Geriatric Society (LKJGG2021W070) and Scientific Research Fund Project of Hebei Provincial Health Commission (20231071).
Author information
Authors and Affiliations
Contributions
LY leads the study design and the overall progress. LZ performed the COPD-related data collection and analysis. ZZ performed the COPD-related data collection and analysis. XJ performed the COPD-related data collection and analysis. TY mainly taken part in the SARS-CoV-2 sequences retrieving and phylogenetic analysis. ZG mainly taken part in the SARS-CoV-2 sequences retrieving and phylogenetic analysis. LY wrote and polished the manuscripts. TY wrote and polished the manuscripts. ZG wrote and polished the manuscripts.
Corresponding author
Ethics declarations
Ethics Approval and Consent to Participate
The study was reviewed and approved by the National Center for Health Statistics Ethics Review Board (Approval Number: 20220611). All authors have their consent to participate.
Consent for Publication
All authors have their consent to publish their work.
Conflict of Interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Zhang, L., Jia, X., Zhang, Z. et al. ceRNA Network Analysis Reveals Potential Key miRNAs and Target Genes in COVID-19-Related Chronic Obstructive Pulmonary Disease. Appl Biochem Biotechnol 196, 4303–4316 (2024). https://doi.org/10.1007/s12010-023-04773-7
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s12010-023-04773-7