Abstract
Purpose of Review
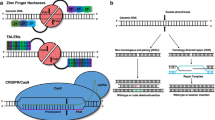
Clustered regularly interspaced short palindromic repeat (CRISPR)/CRISPR-associated 9 (Cas9) has recently emerged as a top genome editing technology and has afforded investigators the ability to more easily study a number of diseases. This review discusses CRISPR/Cas9’s advantages and limitations and highlights a few recent reports on genome editing applications for alleviating dyslipidemia through disruption of proprotein convertase subtilisin/kexin type 9 (PCSK9).
Recent Findings
Targeting of mouse Pcsk9 using CRISPR/Cas9 technology has yielded promising results for lowering total cholesterol levels, and several recent findings are highlighted in this review. Reported on-target mutagenesis efficiency is as high as 90% with a subsequent 40% reduction of blood cholesterol levels in mice, highlighting the potential for use as a therapeutic in human patients.
Summary
The ability to characterize and treat diseases is becoming easier with the recent advances in genome editing technologies. In this review, we discuss how genome editing strategies can be of use for potential therapeutic applications.

Similar content being viewed by others
References
Papers of particular interest, published recently, have been highlighted as: •• Of major importance
Go AS, Mozaffarian D, Roger VL, Benjamin EJ, Berry JD, Borden WB, et al. Heart disease and stroke statistics—2013 update: a report from the American Heart Association. Circulation. 2013;127(1):e6–e245. doi:10.1161/CIR.0b013e31828124ad.
Chadwick AC, Musunuru K. Genome editing for the study of cardiovascular diseases. Curr Cardiol Rep. 2017;19(3):22. doi:10.1007/s11886-017-0830-5.
Strong A, Musunuru K. Genome editing in cardiovascular diseases. Nat Rev Cardiol. 2017;14(1):11–20. doi:10.1038/nrcardio.2016.139.
Seeger T, Porteus M, Wu JC. Genome editing in cardiovascular biology. Circ Res. 2017;120(5):778–80. doi:10.1161/CIRCRESAHA.116.310197.
Termglinchan V, Karakikes I, Seeger T, Wu JC. Current status of genome editing in cardiovascular medicine. In: Turksen K, editor. Genome editing. Cham: Springer International Publishing; 2016. p. 107–26.
Cong L, Ran FA, Cox D, Lin S, Barretto R, Habib N, et al. Multiplex genome engineering using CRISPR/Cas systems. Science. 2013;339(6121):819–23. doi:10.1126/science.1231143.
**ek M, East A, Cheng A, Lin S, Ma E, Doudna J. RNA-programmed genome editing in human cells. elife. 2013;2:e00471. doi:10.7554/eLife.00471.
Cho SW, Kim S, Kim JM, Kim JS. Targeted genome engineering in human cells with the Cas9 RNA-guided endonuclease. Nat Biotechnol. 2013;31(3):230–2. doi:10.1038/nbt.2507.
West SC. Molecular views of recombination proteins and their control. Nat Rev Mol Cell Biol. 2003;4(6):435–45. doi:10.1038/nrm1127.
Hou Z, Zhang Y, Propson NE, Howden SE, Chu LF, Sontheimer EJ, et al. Efficient genome engineering in human pluripotent stem cells using Cas9 from Neisseria meningitidis. Proc Natl Acad Sci U S A. 2013;110(39):15644–9. doi:10.1073/pnas.1313587110.
Esvelt KM, Mali P, Braff JL, Moosburner M, Yaung SJ, Church GM. Orthogonal Cas9 proteins for RNA-guided gene regulation and editing. Nat Methods. 2013;10(11):1116–21. doi:10.1038/nmeth.2681.
•• Ran FA, Cong L, Yan WX, Scott DA, Gootenberg JS, Kriz AJ, et al. In vivo genome editing using Staphylococcus aureus Cas9. Nature. 2015;520(7546):186–91. doi:10.1038/nature14299. This study demonstrated the high efficacy of CRISPR/Cas9 in the mouse liver using Staphylococcus aureus Cas9 delivered by AAV.
Hirano H, Gootenberg JS, Horii T, Abudayyeh OO, Kimura M, Hsu PD, et al. Structure and engineering of Francisella novicida Cas9. Cell. 2016;164(5):950–61. doi:10.1016/j.cell.2016.01.039.
Fu Y, Sander JD, Reyon D, Cascio VM, Joung JK. Improving CRISPR-Cas nuclease specificity using truncated guide RNAs. Nat Biotechnol. 2014;32(3):279–84. doi:10.1038/nbt.2808.
Hsu PD, Scott DA, Weinstein JA, Ran FA, Konermann S, Agarwala V, et al. DNA targeting specificity of RNA-guided Cas9 nucleases. Nat Biotechnol. 2013;31(9):827–32. doi:10.1038/nbt.2647.
Ran FA, Hsu PD, Lin CY, Gootenberg JS, Konermann S, Trevino AE, et al. Double nicking by RNA-guided CRISPR Cas9 for enhanced genome editing specificity. Cell. 2013;154(6):1380–9. doi:10.1016/j.cell.2013.08.021.
Mali P, Aach J, Stranges PB, Esvelt KM, Moosburner M, Kosuri S, et al. CAS9 transcriptional activators for target specificity screening and paired nickases for cooperative genome engineering. Nat Biotechnol. 2013;31(9):833–8. doi:10.1038/nbt.2675.
Guilinger JP, Thompson DB, Liu DR. Fusion of catalytically inactive Cas9 to FokI nuclease improves the specificity of genome modification. Nat Biotechnol. 2014;32(6):577–82. doi:10.1038/nbt.2909.
Slaymaker IM, Gao L, Zetsche B, Scott DA, Yan WX, Zhang F. Rationally engineered Cas9 nucleases with improved specificity. Science. 2016;351(6268):84–8. doi:10.1126/science.aad5227.
Kleinstiver BP, Pattanayak V, Prew MS, Tsai SQ, Nguyen NT, Zheng Z, et al. High-fidelity CRISPR-Cas9 nucleases with no detectable genome-wide off-target effects. Nature. 2016;529(7587):490–5. doi:10.1038/nature16526.
Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533(7603):420–4. doi:10.1038/nature17946.
Ma Y, Zhang J, Yin W, Zhang Z, Song Y, Chang X. Targeted AID-mediated mutagenesis (TAM) enables efficient genomic diversification in mammalian cells. Nat Methods. 2016;13(12):1029–35. doi:10.1038/nmeth.4027.
Hess GT, Fresard L, Han K, Lee CH, Li A, Cimprich KA, et al. Directed evolution using dCas9-targeted somatic hypermutation in mammalian cells. Nat Methods. 2016;13(12):1036–42. doi:10.1038/nmeth.4038.
Nishida K, Arazoe T, Yachie N, Banno S, Kakimoto M, Tabata M, et al. Targeted nucleotide editing using hybrid prokaryotic and vertebrate adaptive immune systems. Science. 2016;16:353(6305).
Kleinstiver BP, Prew MS, Tsai SQ, Topkar VV, Nguyen NT, Zheng Z, et al. Engineered CRISPR-Cas9 nucleases with altered PAM specificities. Nature. 2015;523(7561):481–5. doi:10.1038/nature14592.
Zhang Y, Ge X, Yang F, Zhang L, Zheng J, Tan X, et al. Comparison of non-canonical PAMs for CRISPR/Cas9-mediated DNA cleavage in human cells. Sci Rep. 2014;4:5405. doi:10.1038/srep05405.
Ding Q, Lee YK, Schaefer EA, Peters DT, Veres A, Kim K, et al. A TALEN genome-editing system for generating human stem cell-based disease models. Cell Stem Cell. 2013;12(2):238–51. doi:10.1016/j.stem.2012.11.011.
Gupta RM, Meissner TB, Cowan CA, Musunuru K. Genome-edited human pluripotent stem cell-derived macrophages as a model of reverse cholesterol transport—brief report. Arterioscler Thromb Vasc Biol. 2016;36(1):15–8. doi:10.1161/ATVBAHA.115.305956.
Pashos EE, Park Y, Wang X, Raghavan A, Yang W, Abbey D, et al. Large, diverse population cohorts of hiPSCs and derived hepatocyte-like cells reveal functional genetic variation at blood lipid-associated loci. Cell Stem Cell. 2017;20(4):558–570 e10. doi:10.1016/j.stem.2017.03.017.
Maetzel D, Sarkar S, Wang H, Abi-Mosleh L, Xu P, Cheng AW, et al. Genetic and chemical correction of cholesterol accumulation and impaired autophagy in hepatic and neural cells derived from Niemann-Pick Type C patient-specific iPS cells. Stem Cell Reports. 2014;2(6):866–80. doi:10.1016/j.stemcr.2014.03.014.
Cayo MA, Mallanna SK, Di Furio F, **g R, Tolliver LB, Bures M, et al. A drug screen using human iPSC-derived hepatocyte-like cells reveals cardiac glycosides as a potential treatment for hypercholesterolemia. Cell Stem Cell. 2017;20(4):478–489 e5. doi:10.1016/j.stem.2017.01.011.
Warren CR, O'Sullivan JF, Friesen M, Becker CE, Zhang X, Liu P, et al. Induced pluripotent stem cell differentiation enables functional validation of GWAS variants in metabolic disease. Cell Stem Cell. 2017;20(4):547–557 e7. doi:10.1016/j.stem.2017.01.010.
Wang H, Yang H, Shivalila CS, Dawlaty MM, Cheng AW, Zhang F, et al. One-step generation of mice carrying mutations in multiple genes by CRISPR/Cas-mediated genome engineering. Cell. 2013;153(4):910–8. doi:10.1016/j.cell.2013.04.025.
Yang H, Wang H, Shivalila CS, Cheng AW, Shi L, Jaenisch R. One-step generation of mice carrying reporter and conditional alleles by CRISPR/Cas-mediated genome engineering. Cell. 2013;154(6):1370–9. doi:10.1016/j.cell.2013.08.022.
•• Ding Q, Strong A, Patel KM, Ng SL, Gosis BS, Regan SN, et al. Permanent alteration of PCSK9 with in vivo CRISPR-Cas9 genome editing. Circ Res. 2014;115(5):488–92. doi:10.1161/CIRCRESAHA.115.304351. This study established the high efficacy of CRISPR/Cas9 targeting of Pcsk9 in the mouse liver.
•• Wang X, Raghavan A, Chen T, Qiao L, Zhang Y, Ding Q, et al. CRISPR-Cas9 targeting of PCSK9 in human hepatocytes in vivo—brief report. Arterioscler Thromb Vasc Biol. 2016;36(5):783–6. doi:10.1161/ATVBAHA.116.307227. This study demonstrated high efficacy of CRISPR/Cas9 in a liver-humanized mouse model.
Jacob HJ, Lazar J, Dwinell MR, Moreno C, Geurts AM. Gene targeting in the rat: advances and opportunities. Trends Genet. 2010;26(12):510–8. doi:10.1016/j.tig.2010.08.006.
Moreno C, Hoffman M, Stodola TJ, Didier DN, Lazar J, Geurts AM, et al. Creation and characterization of a renin knockout rat. Hypertension. 2011;57(3):614–9. doi:10.1161/HYPERTENSIONAHA.110.163840.
Endres BT, Priestley JR, Palygin O, Flister MJ, Hoffman MJ, Weinberg BD, et al. Mutation of Plekha7 attenuates salt-sensitive hypertension in the rat. Proc Natl Acad Sci U S A. 2014;111(35):12817–22. doi:10.1073/pnas.1410745111.
Larcher T, Lafoux A, Tesson L, Remy S, Thepenier V, Francois V, et al. Characterization of dystrophin deficient rats: a new model for Duchenne muscular dystrophy. PLoS One. 2014;9(10):e110371. doi:10.1371/journal.pone.0110371.
Li J, **e D, Huang J, Lv F, Shi D, Liu Y, et al. Cold-inducible RNA-binding protein regulates cardiac repolarization by targeting transient outward potassium channels. Circ Res. 2015;116(10):1655–9. doi:10.1161/CIRCRESAHA.116.306287.
Zhu X, Fang J, Jiang DS, Zhang P, Zhao GN, Zhu X, et al. Exacerbating pressure overload-induced cardiac hypertrophy: novel role of adaptor molecule Src homology 2-B3. Hypertension. 2015;66(3):571–81. doi:10.1161/HYPERTENSIONAHA.115.05183.
Yang D, Xu J, Zhu T, Fan J, Lai L, Zhang J, et al. Effective gene targeting in rabbits using RNA-guided Cas9 nucleases. J Mol Cell Biol. 2014;6(1):97–9. doi:10.1093/jmcb/mjt047.
Ji D, Zhao G, Songstad A, Cui X, Weinstein EJ. Efficient creation of an APOE knockout rabbit. Transgenic Res. 2015;24(2):227–35. doi:10.1007/s11248-014-9834-8.
Niimi M, Yang D, Kitajima S, Ning B, Wang C, Li S, et al. ApoE knockout rabbits: a novel model for the study of human hyperlipidemia. Atherosclerosis. 2016;245:187–93. doi:10.1016/j.atherosclerosis.2015.12.002.
Zou J, Tran D, Baalbaki M, Tang LF, Poon A, Pelonero A, et al. An internal promoter underlies the difference in disease severity between N- and C-terminal truncation mutations of Titin in zebrafish. elife. 2015;4:e09406. doi:10.7554/eLife.09406.
Wang Y, Du Y, Shen B, Zhou X, Li J, Liu Y, et al. Efficient generation of gene-modified pigs via injection of zygote with Cas9/sgRNA. Sci Rep. 2015;5:8256. doi:10.1038/srep08256.
Umeyama K, Watanabe K, Watanabe M, Horiuchi K, Nakano K, Kitashiro M, et al. Generation of heterozygous fibrillin-1 mutant cloned pigs from genome-edited foetal fibroblasts. Sci Rep. 2016;6:24413. doi:10.1038/srep24413.
Cohen JC, Boerwinkle E, Mosley TH Jr, Hobbs HH. Sequence variations in PCSK9, low LDL, and protection against coronary heart disease. N Engl J Med. 2006;354(12):1264–72. doi:10.1056/NEJMoa054013.
Sabatine MS, Giugliano RP, Keech AC, Honarpour N, Wiviott SD, Murphy SA, et al. Evolocumab and clinical outcomes in patients with cardiovascular disease. N Engl J Med. 2017; doi:10.1056/NEJMoa1615664.
Muruve DA. The innate immune response to adenovirus vectors. Hum Gene Ther. 2004;15(12):1157–66. doi:10.1089/hum.2004.15.1157.
Gregory SM, Nazir SA, Metcalf JP. Implications of the innate immune response to adenovirus and adenoviral vectors. Future Virol. 2011;6(3):357–74. doi:10.2217/fvl.11.6.
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of Interest
Alexandra Chadwick and Kiran Musunuru declare they have no conflicts of interest.
Human and Animal Rights and Informed Consent
All procedures performed in studies involving human participants were in accordance with the ethical standards of the institutional and/or national research committee and with the 1964 Helsinki declaration and its later amendments or comparable ethical standards. All applicable international, national, and/or institutional guidelines for the care and use of animals were followed. All procedures performed in studies involving animals were in accordance with the ethical standards of the institution or practice at which the studies were conducted.
Additional information
This article is part of the Topical Collection on Genetics and Genomics
Rights and permissions
About this article
Cite this article
Chadwick, A.C., Musunuru, K. Treatment of Dyslipidemia Using CRISPR/Cas9 Genome Editing. Curr Atheroscler Rep 19, 32 (2017). https://doi.org/10.1007/s11883-017-0668-8
Published:
DOI: https://doi.org/10.1007/s11883-017-0668-8