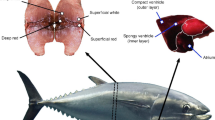
Abstract
Although most fishes are ectothermic, some, including tuna and billfish, achieve endothermy through specialized heat producing tissues that are modified muscles. How these heat producing tissues evolved, and whether they share convergent molecular mechanisms, remain unresolved. Here, we generated a high-quality genome from the mackerel tuna (Euthynnus affinis) and investigated the heat producing tissues of this fish by single-nucleus and bulk RNA sequencing. Compared with other teleosts, tuna-specific genetic variation is strongly associated with muscle differentiation. Single-nucleus RNA-seq revealed a high proportion of specific slow skeletal muscle cell subtypes in the heat producing tissues of tuna. Marker genes of this cell subtype are associated with the relative sliding of actin and myosin, suggesting that tuna endothermy is mainly based on shivering thermogenesis. In contrast, cross-species transcriptome analysis indicated that endothermy in billfish relies mainly on non-shivering thermogenesis. Nevertheless, the heat producing tissues of the different species do share some tissue-specific genes, including vascular-related and mitochondrial genes. Overall, although tunas and billfishes differ in their thermogenic strategies, they share similar expression patterns in some respects, highlighting the complexity of convergent evolution.
Similar content being viewed by others
References
Allen, J.E., Majoros, W.H., Pertea, M., and Salzberg, S.L. (2006). JIGSAW, GeneZilla, and GlimmerHMM: puzzling out the features of human genes in the ENCODE regions. Genome Biol 7, S9.
Bal, N.C., Maurya, S.K., Sopariwala, D.H., Sahoo, S.K., Gupta, S.C., Shaikh, S.A., Pant, M., Rowland, L.A., Bombardier, E., Goonasekera, S.A., et al. (2012). Sarcolipin is a newly identified regulator of muscle-based thermogenesis in mammals. Nat Med 18, 1575–1579.
Benson, G. (1999). Tandem repeats finder: a program to analyze DNA sequences. Nucleic Acids Res 27, 573–580.
Birney, E., Clamp, M., and Durbin, R. (2004). GeneWise and genomewise. Genome Res 14, 988–995.
Blanchette, M., Kent, W.J., Riemer, C., Elnitski, L., Smit, A.F.A., Roskin, K.M., Baertsch, R., Rosenbloom, K., Clawson, H., Green, E.D., et al. (2004). Aligning multiple genomic sequences with the threaded blockset aligner. Genome Res 14, 708–715.
Block, B.A. (1986). Structure of the brain and eye heater tissue in marlins, sailfish, and spearfishes. J Morphol 190, 169–189.
Block, B.A. (1991). Evolutionary novelties: How fish have built a heater out of muscle. Am Zool 31, 726–742.
Boye, J., Musyl, M., Brill, R., and Malte, H. (2009). Transectional heat transfer in thermoregulating bigeye tuna (Thunnus obesus)—a 2D heat flux model. J Exp Biol 212, 3708–3718.
Chen, B., Xu, X., Lin, D., Chen, X., Xu, Y., Liu, X., and Dong, W. (2021a). KRT18 modulates alternative splicing of genes involved in proliferation and apoptosis processes in both gastric cancer cells and clinical samples. Front Genet 12, 635429.
Chen, S., Zhou, Y., Chen, Y., and Gu, J. (2018). fastp: an ultra-fast all-in-one FASTQ preprocessor. Bioinformatics 34, i884–i890.
Chen, Z., Zhang, H., Bai, Y., Cui, C., Li, S., Wang, W., Deng, Y., Gao, Q., Wang, L., Qi, W., et al. (2021b). Single cell transcriptomic analysis identifies novel vascular smooth muscle subsets under high hydrostatic pressure. Sci China Life Sci 64, 1677–1690.
Cheng, H., Concepcion, G.T., Feng, X., Zhang, H., and Li, H. (2021). Haplotype-resolved de novo assembly using phased assembly graphs with hifiasm. Nat Methods 18, 170–175.
Ciezarek, A., Gardner, L., Savolainen, V., and Block, B. (2020). Skeletal muscle and cardiac transcriptomics of a regionally endothermic fish, the Pacific bluefin tuna, Thunnus orientalis. BMC Genomics 21, 642.
Cudmore, M.J., Hewett, P.W., Ahmad, S., Wang, K.Q., Cai, M., Al-Ani, B., Fujisawa, T., Ma, B., Sissaoui, S., Ramma, W., et al. (2012). The role of heterodimerization between VEGFR-1 and VEGFR-2 in the regulation of endothelial cell homeostasis. Nat Commun 3, 972.
Davenport, J., Jones, T.T., Work, T.M., and Balazs, G.H. (2015). Topsyturvy: turning the counter-current heat exchange of leatherback turtles upside down. Biol Lett 11, 20150592.
De Bie, T., Cristianini, N., Demuth, J.P., and Hahn, M.W. (2006). CAFE: a computational tool for the study of gene family evolution. Bioinformatics 22, 1269–1271.
Dickson, K.A. (1995). Unique adaptations of the metabolic biochemistry of tunas and billfishes for life in the pelagic environment. Environ Biol Fish 42, 65–97.
Dickson, K.A., and Graham, J.B. (2004). Evolution and consequences of endothermy in fishes. Physiol Biochem Zool 77, 998–1018.
Dudchenko, O., Batra, S.S., Omer, A.D., Nyquist, S.K., Hoeger, M., Durand, N.C., Shamim, M.S., Machol, I., Lander, E.S., Aiden, A.P., et al. (2017). De novo assembly of the Aedes aegypti genome using Hi-C yields chromosome-length scaffolds. Science 356, 92–95.
Durand, N.C., Robinson, J.T., Shamim, M.S., Machol, I., Mesirov, J.P., Lander, E.S., and Aiden, E.L. (2016a). Juicebox provides a visualization system for Hi-C contact maps with unlimited zoom. Cell Syst 3, 99–101.
Durand, N.C., Shamim, M.S., Machol, I., Rao, S.S.P., Huntley, M.H., Lander, E.S., and Aiden, E.L. (2016b). Juicer provides a one-click system for analyzing loop-resolution Hi-C experiments. Cell Syst 3, 95–98.
Emms, D.M., and Kelly, S. (2015). OrthoFinder: solving fundamental biases in whole genome comparisons dramatically improves orthogroup inference accuracy. Genome Biol 16, 157.
Foster, C.S.P., Van Dyke, J.U., Thompson, M.B., Smith, N.M.A., Simpfendorfer, C.A., Murphy, C.R., and Whittington, C.M. (2022). Different genes are recruited during convergent evolution of pregnancy and the placenta. Mol Biol Evol 39.
Gallant, J.R., Traeger, L.L., Volkening, J.D., Moffett, H., Chen, P.H., Novina, C.D., Phillips Jr., G.N., Anand, R., Wells, G.B., Pinch, M., et al. (2014). Genomic basis for the convergent evolution of electric organs. Science 344, 1522–1525.
Gómez-Salinero, J.M., Izzo, F., Lin, Y., Houghton, S., Itkin, T., Geng, F., Bram, Y., Adelson, R.P., Lu, T.M., Inghirami, G., et al. (2022). Specification of fetal liver endothelial progenitors to functional zonated adult sinusoids requires c-Maf induction. Cell Stem Cell 29, 593–609.e7.
Graham, J.B., Koehrn, F.J., and Dickson, K.A. (1983). Distribution and relative proportions of red muscle in scombrid fishes: consequences of body size and relationships to locomotion and endothermy. Can J Zool 61, 2087–2096.
Grant, T.R., and Dawson, T.J. (1978). Temperature regulation in the platypus, ornithorhynchus anatinus: production and loss of metabolic heat in air and water. Physiol Zool 51, 315–332.
Haas, B.J., Salzberg, S.L., Zhu, W., Pertea, M., Allen, J.E., Orvis, J., White, O., Buell, C.R., and Wortman, J.R. (2008). Automated eukaryotic gene structure annotation using evidencemodeler and the program to assemble spliced alignments. Genome Biol 9, R7.
Han, L., Wei, X., Liu, C., Volpe, G., Zhuang, Z., Zou, X., Wang, Z., Pan, T., Yuan, Y., Zhang, X., et al. (2022). Cell transcriptomic atlas of the non-human primate Macaca fascicularis. Nature 604, 723–731.
Hastings, K.E.M. (1997). Molecular evolution of the vertebrate troponin I gene family. Cell Struct Funct 22, 205–211.
Havelka, M., Sawayama, E., Saito, T., Yoshitake, K., Saka, D., Ineno, T., Asakawa, S., Takagi, M., Goto, R., and Matsubara, T. (2021). Chromosome-scale genome assembly and transcriptome assembly of kawakawa Euthynnus affinis a tuna-like species. Front Genet 12, 739781.
Hernández-García, R., Iruela-Arispe, M.L., Reyes-Cruz, G., and Vázquez-Prado, J. (2015). Endothelial RhoGEFs: A systematic analysis of their expression profiles in VEGF-stimulated and tumor endothelial cells. Vascul Pharmacol 74, 60–72.
Hernandez, O.M., Jones, M., Guzman, G., and Szczesna-Cordary, D. (2007). Myosin essential light chain in health and disease. Am J Physiol Heart Circ Physiol 292, H1643–H1654.
Hu, M.L., Wu, B.S., Xu, W.J., Zhu, C.L., Yuan, Y., Lv, W.Q., Qiu, Q., Zhang, H.B., Wang, K., and Feng, C.G. (2022). Gene expression responses in zebrafish to short-term high-hydrostatic pressure. Zool Res 43, 188–191.
Jones, P., Binns, D., Chang, H.Y., Fraser, M., Li, W., McAnulla, C., McWilliam, H., Maslen, J., Mitchell, A., Nuka, G., et al. (2014). InterProScan 5: genome-scale protein function classification. Bioinformatics 30, 1236–1240.
Kanehisa, M., and Goto, S. (2000). KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res 28, 27–30.
Kedlian, V.R., Wang, Y., Liu, T., Chen, X., Bolt, L., Shen, Z., Fasouli, E.S., Prigmore, E., Kleshchevnikov, V., Li, T., et al. (2022). Human skeletal muscle ageing atlas. bioRxiv, 2022.2005.2024.493094.
Kielbasa, S.M., Wan, R., Sato, K., Horton, P., and Frith, M.C. (2011). Adaptive seeds tame genomic sequence comparison. Genome Res 21, 487–493.
Legendre, L.J., and Davesne, D. (2020). The evolution of mechanisms involved in vertebrate endothermy. Phil Trans R Soc B 375, 20190136.
Leinwand, L.A., Saez, L., McNally, E., and Nadal-Ginard, B. (1983). Isolation and characterization of human myosin heavy chain genes. Proc Natl Acad Sci USA 80, 3716–3720.
Li, H., and Durbin, R. (2009). Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760.
Li, H., Handsaker, B., Wysoker, A., Fennell, T., Ruan, J., Homer, N., Marth, G., Abecasis, G., and Durbin, R. (2009). The sequence alignment/map format and SAMtools. Bioinformatics 25, 2078–2079.
Luna, V.M., Daikoku, E., and Ono, F. (2015). “Slow” skeletal muscles across vertebrate species. Cell Biosci 5, 62.
Luo, R., Liu, B., **e, Y., Li, Z., Huang, W., Yuan, J., He, G., Chen, Y., Pan, Q., Liu, Y., et al. (2012). SOAPdenovo2: an empirically improved memory-efficient short-read de novo assembler. GigaScience 1, 18.
Madigan, D.J., Carlisle, A.B., Gardner, L.D., Jayasundara, N., Micheli, F., Schaefer, K.M., Fuller, D.W., and Block, B.A. (2015). Assessing niche width of endothermic fish from genes to ecosystem. Proc Natl Acad Sci USA 112, 8350–8355.
Manni, M., Berkeley, M.R., Seppey, M., Simão, F.A., and Zdobnov, E.M. (2021). BUSCO update: novel and streamlined workflows along with broader and deeper phylogenetic coverage for scoring of eukaryotic, prokaryotic, and viral genomes. Mol Biol Evol 38, 4647–4654.
Marcovitz, A., Turakhia, Y., Chen, H.I., Gloudemans, M., Braun, B.A., Wang, H., and Bejerano, G. (2019). A functional enrichment test for molecular convergent evolution finds a clear protein-coding signal in echolocating bats and whales. Proc Natl Acad Sci USA 116, 21094–21103.
Meredith, C., Herrmann, R., Parry, C., Liyanage, K., Dye, D.E., Durling, H. J., Duff, R.M., Beckman, K., de Visser, M., van der Graaff, M.M., et al. (2004). Mutations in the slow skeletal muscle fiber myosin heavy chain gene (MYH7) cause laing early-onset distal myopathy (MPD1). Am J Hum Genet 75, 703–708.
Metikala, S., Casie Chetty, S., and Sumanas, S. (2021). Single-cell transcriptome analysis of the zebrafish embryonic trunk. PLoS ONE 16, e0254024.
Mueller, S.B., and Kontos, C.D. (2016). Tie1: an orphan receptor provides context for angiopoietin-2/Tie2 signaling. J Clin Invest 126, 3188–3191.
Nakamura, Y., Mori, K., Saitoh, K., Oshima, K., Mekuchi, M., Sugaya, T., Shigenobu, Y., Ojima, N., Muta, S., Fujiwara, A., et al. (2013). Evolutionary changes of multiple visual pigment genes in the complete genome of Pacific bluefin tuna. Proc Natl Acad Sci USA 110, 11061–11066.
Nesmith, J.E., Chappell, J.C., Cluceru, J.G., and Bautch, V.L. (2017). Blood vessel anastomosis is spatially regulated by Flt1 during angiogenesis. Development 144, 889–896.
Noviski, M., Mueller, J.L., Satterthwaite, A., Garrett-Sinha, L.A., Brombacher, F., and Zikherman, J. (2018). IgM and IgD B cell receptors differentially respond to endogenous antigens and control B cell fate. Elife 7, e35074.
Nowack, J., Giroud, S., Arnold, W., and Ruf, T. (2017). Muscle non-shivering thermogenesis and its role in the evolution of endothermy. Front Physiol 8, 889.
Oldfors, A. (2007). Hereditary myosin myopathies. Neuromuscul Disord 17, 355–367.
Periasamy, M., Herrera, J.L., and Reis, F.C.G. (2017). Skeletal muscle thermogenesis and its role in whole body energy metabolism. Diabetes Metab J 41, 327–336.
Pertea, M., Pertea, G.M., Antonescu, C.M., Chang, T.C., Mendell, J.T., and Salzberg, S.L. (2015). StringTie enables improved reconstruction of a transcriptome from RNA-seq reads. Nat Biotechnol 33, 290–295.
Rowland, L.A., Bal, N.C., and Periasamy, M. (2015). The role of skeletal-muscle-based thermogenic mechanisms in vertebrate endothermy. Biol Rev 90, 1279–1297.
Rueden, C.T., Schindelin, J., Hiner, M.C., DeZonia, B.E., Walter, A.E., Arena, E.T., and Eliceiri, K.W. (2017). ImageJ2: ImageJ for the next generation of scientific image data. BMC BioInf 18, 529.
Saha, S., Bridges, S., Magbanua, Z.V., and Peterson, D.G. (2008). Empirical comparison of ab initio repeat finding programs. Nucleic Acids Res 36, 2284–2294.
Shafer, M.E.R., Sawh, A.N., and Schier, A.F. (2022). Gene family evolution underlies cell-type diversification in the hypothalamus of teleosts. Nat Ecol Evol 6, 63–76.
Shelton, M., Ritso, M., Liu, J., O’Neil, D., Kocharyan, A., Rudnicki, M.A., Stanford, W.L., Skerjanc, I.S., and Blais, A. (2019). Gene expression profiling of skeletal myogenesis in human embryonic stem cells reveals a potential cascade of transcription factors regulating stages of myogenesis, including quiescent/activated satellite cell-like gene expression. PLoS ONE 14, e0222946.
Sheng, J.J., and **, J.P. (2016). TNNI1, TNNI2 and TNNI3 evolution, regulation, and protein structure-function relationships. Gene 576, 385–394.
Stamatakis, A. (2014). RAxML version 8: a tool for phylogenetic analysis and post-analysis of large phylogenies. Bioinformatics 30, 1312–1313.
Stanke, M., and Morgenstern, B. (2005). AUGUSTUS: a web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res 33, W465–W467.
Stoehr, A., Martin, J.S., Aalbers, S., Sepulveda, C., and Bernal, D. (2018). Free-swimming swordfish, **phias gladius, alter the rate of whole body heat transfer: morphological and physiological specializations for thermoregulation. ICES J Mar Sci 75, 858–870.
Syme, D.A., and Shadwick, R.E. (2011). Red muscle function in stiff-bodied swimmers: there and almost back again. Phil Trans R Soc B 366, 1507–1515.
Tarailo-Graovac, M., and Chen, N. (2009). Using RepeatMasker to identify repetitive elements in genomic sequences. Curr Protoc Bioinformatics 25, 4–10.
Taylor, M.V., and Hughes, S.M. (2017). Mef2 and the skeletal muscle differentiation program. Semin Cell Dev Biol 72, 33–44.
Tullis, A., and Block, B.A. (1997). Histochemical and immunohistochemical studies on the origin of the blue marlin heater cell phenotype. Tissue Cell 29, 627–642.
Vestweber, D. (2021). Vascular endothelial protein tyrosine phosphatase regulates endothelial function. Physiology 36, 84–93.
Wang, M., Zhao, Y., and Zhang, B. (2015). Efficient test and visualization of multi-set intersections. Sci Rep 5, 16923.
Wang, X., Qu, M., Liu, Y., Schneider, R.F., Song, Y., Chen, Z., Zhang, H., Zhang, Y., Yu, H., Zhang, S., et al. (2022). Genomic basis of evolutionary adaptation in a warm-blooded fish. Innovation 3, 100185.
Watanabe, Y.Y., Goldman, K.J., Caselle, J.E., Chapman, D.D., and Papastamatiou, Y.P. (2015). Comparative analyses of animal-tracking data reveal ecological significance of endothermy in fishes. Proc Natl Acad Sci USA 112, 6104–6109.
Wei, B., and **, J.P. (2016). TNNT1, TNNT2, and TNNT3 isoform genes, regulation, and structure-function relationships. Gene 582, 1–13.
Weiss, A., McDonough, D., Wertman, B., Acakpo-Satchivi, L., Montgomery, K., Kucherlapati, R., Leinwand, L., and Krauter, K. (1999). Organization of human and mouse skeletal myosin heavy chain gene clusters is highly conserved. Proc Natl Acad Sci USA 96, 2958–2963.
Wolf, F.A., Angerer, P., and Theis, F.J. (2018). SCANPY: large-scale single-cell gene expression data analysis. Genome Biol 19, 15.
Wolock, S.L., Lopez, R., and Klein, A.M. (2019). Scrublet: computational identification of cell doublets in single-cell transcriptomic data. Cell Syst 8, 281–291.e9.
Wu, B., Feng, C., Zhu, C., Xu, W., Yuan, Y., Hu, M., Yuan, K., Li, Y., Ren, Y., Zhou, Y., et al. (2021). The genomes of two billfishes provide insights into the evolution of endothermy in teleosts. Mol Biol Evol 38, 2413–2427.
Yoshida, M.A., Ogura, A., Ikeo, K., Shigeno, S., Moritaki, T., Winters, G. C., Kohn, A.B., and Moroz, L.L. (2015). Molecular evidence for convergence and parallelism in evolution of complex brains of cephalopod Molluscs: Insights from visual systems. Integr Comp Biol 55, 1070–1083.
Young, M.D., and Behjati, S. (2020). SoupX removes ambient RNA contamination from droplet-based single-cell RNA sequencing data. Gigascience 9.
Zhang, Y., Park, C., Bennett, C., Thornton, M., and Kim, D. (2021). Rapid and accurate alignment of nucleotide conversion sequencing reads with HISAT-3N. Genome Res 31, 1290–1295.
Acknowledgements
This work was supported by the National Natural Science Foundation of China (32122021), the National Key Research and Development Program of China (2022YFC3400300), the 1000 Talent Project of Shaanxi Province to Q.Q. and K.W., the Fundamental Research Funds of Northwestern Polytechnic University, and the Open Foundation from Marine Sciences in the First-Class Subjects of Zhejiang (OFMS011). We thank Zhengyi Fu, Bo ** He
Corresponding authors
Ethics declarations
The author(s) declare that they have no conflict of interest.
Supporting Information
Rights and permissions
About this article
Cite this article
Wu, B., Gao, X., Hu, M. et al. Distinct and shared endothermic strategies in the heat producing tissues of tuna and other teleosts. Sci. China Life Sci. 66, 2629–2645 (2023). https://doi.org/10.1007/s11427-022-2312-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11427-022-2312-1