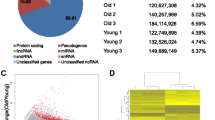
Abstract
The liver, as a crucial metabolic organ, undergoes significant pathological changes during the aging process, which can have a profound impact on overall health. To gain a comprehensive understanding of these alterations, we employed data-driven approaches, along with biochemical methods, histology, and immunohistochemistry techniques, to systematically investigate the effects of aging on the liver. Our study utilized a well-established rat aging model provided by the National Institute of Aging. Systems biology approaches were used to analyze genome-wide transcriptomics data from liver samples obtained from young (4–5 months old) and aging (20–21 months old) Fischer 344 rats. Our findings revealed pathological changes occurring in various essential biological processes in aging livers. These included mitochondrial dysfunction, increased oxidative/nitrative stress, decreased NAD + content, impaired amino acid and protein synthesis, heightened inflammation, disrupted lipid metabolism, enhanced apoptosis, senescence, and fibrosis. These results were validated using independent datasets from both human and rat aging studies. Furthermore, by employing co-expression network analysis, we identified novel driver genes responsible for liver aging, confirmed our findings in human aging subjects, and pointed out the cellular localization of the driver genes using single-cell RNA-sequencing human data. Our study led to the discovery and validation of a liver-specific gene, proprotein convertase subtilisin/kexin type 9 (PCSK9), as a potential therapeutic target for mitigating the pathological processes associated with aging in the liver. This finding envisions new possibilities for develo** interventions aimed to improve liver health during the aging process.










Similar content being viewed by others
References
United Nations. World Population Ageing 2020 Highlights. 2020. https://www.un.org/development/desa/pd/sites/www.un.org.development.desa.pd/files/files/documents/2020/Sep/un_pop_2020_pf_ageing_10_key_messages.pdf. Accessed 10 Nov 2022.
WHO. Ageing and health. 2021. https://www.who.int/news-room/fact-sheets/detail/ageing-and-health. Accessed 10 Nov 2022.
Lopez-Otin C, Blasco MA, Partridge L, Serrano M, Kroemer G. The hallmarks of aging. Cell. 2013;153:1194–217.
Schmucker DL. Age-related changes in liver structure and function: Implications for disease? Exp Gerontol. 2005;40:650–9.
Matyas C, Hasko G, Liaudet L, Trojnar E, Pacher P. Interplay of cardiovascular mediators, oxidative stress and inflammation in liver disease and its complications. Nat Rev Cardiol. 2021;18:117–35.
Kim IH, Kisseleva T, Brenner DA. Aging and liver disease. Curr Opin Gastroenterol. 2015;31:184–91.
Shavlakadze T, Morris M, Fang J, Wang SX, Zhu J, Zhou W, Tse HW, Mondragon-Gonzalez R, Roma G, Glass DJ. Age-related gene expression signature in rats demonstrate early, late, and linear transcriptional changes from multiple tissues. Cell Rep. 2019;28:3263-3273 e3263.
Shavlakadze T, Zhu J, Wang S, Zhou W, Morin B, Egerman MA, Fan L, Wang Y, Iartchouk O, Meyer A, Valdez RA, Mannick JB, Klickstein LB, Glass DJ. Short-term low-dose mTORC1 inhibition in aged rats counter-regulates age-related gene changes and blocks age-related kidney pathology. J Gerontol A Biol Sci Med Sci. 2018;73:845–52.
Ding J, Ji J, Rabow Z, Shen T, Folz J, Brydges CR, Fan S, Lu X, Mehta S, Showalter MR, Zhang Y, Araiza R, Bower LR, Lloyd KCK, Fiehn O. A metabolome atlas of the aging mouse brain. Nat Commun. 2021;12:6021.
**e K, Qin Q, Long Z, Yang Y, Peng C, ** C, Li L, Wu Z, Daria V, Zhao Y, Wang F, Wang M. High-throughput metabolomics for discovering potential biomarkers and identifying metabolic mechanisms in aging and Alzheimer’s Disease. Front Cell Dev Biol. 2021;9:602887.
Tanaka T, Basisty N, Fantoni G, Candia J, Moore AZ, Biancotto A, Schilling B, Bandinelli S, Ferrucci L. Plasma proteomic biomarker signature of age predicts health and life span. Elife. 2020;9:e61073.
Zhang C, Bjornson E, Arif M, Tebani A, Lovric A, Benfeitas R, Ozcan M, Juszczak K, Kim W, Kim JT, Bidkhori G, Stahlman M, Bergh PO, Adiels M, Turkez H, Taskinen MR, Bosley J, Marschall HU, Nielsen J, Uhlen M, Boren J, Mardinoglu A. The acute effect of metabolic cofactor supplementation: a potential therapeutic strategy against non-alcoholic fatty liver disease. Mol Syst Biol. 2020;16:e9495.
Turanli B, Zhang C, Kim W, Benfeitas R, Uhlen M, Arga KY, Mardinoglu A. Discovery of therapeutic agents for prostate cancer using genome-scale metabolic modeling and drug repositioning. EBioMedicine. 2019;42:386–96.
Benfeitas R, Bidkhori G, Mukhopadhyay B, Klevstig M, Arif M, Zhang C, Lee S, Cinar R, Nielsen J, Uhlen M, Boren J, Kunos G, Mardinoglu A. Characterization of heterogeneous redox responses in hepatocellular carcinoma patients using network analysis. EBioMedicine. 2019;40:471–87.
Arif M, Zhang C, Li X, Gungor C, Cakmak B, Arslanturk M, Tebani A, Ozcan B, Subas O, Zhou W, Piening B, Turkez H, Fagerberg L, Price N, Hood L, Snyder M, Nielsen J, Uhlen M, Mardinoglu A. iNetModels 2.0: an interactive visualization and database of multi-omics data. Nucleic Acids Res. 2021;49:W271–6.
Zeybel M, Arif M, Li X, Altay O, Yang H, Shi M, Akyildiz M, Saglam B, Gonenli MG, Yigit B, Ulukan B, Ural D, Shoaie S, Turkez H, Nielsen J, Zhang C, Uhlen M, Boren J, Mardinoglu A. Multiomics analysis reveals the impact of microbiota on host metabolism in hepatic steatosis. Adv Sci (Weinh). 2022;9:e2104373.
Arif M, Klevstig M, Benfeitas R, Doran S, Turkez H, Uhlen M, Clausen M, Wikstrom J, Etal D, Zhang C, Levin M, Mardinoglu A, Boren J. Integrative transcriptomic analysis of tissue-specific metabolic crosstalk after myocardial infarction. Elife. 2021;10.
Gene Ontology C. The gene ontology resource: enriching a GOld mine. Nucleic Acids Res. 2021;49:D325–34.
Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat Genet. 2000;25:25–9.
Kanehisa M, Furumichi M, Sato Y, Ishiguro-Watanabe M, Tanabe M. KEGG: integrating viruses and cellular organisms. Nucleic Acids Res. 2021;49:D545–51.
Kanehisa M. Toward understanding the origin and evolution of cellular organisms. Protein Sci. 2019;28:1947–51.
Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30.
Herrera J, Henke CA, Bitterman PB. Extracellular matrix as a driver of progressive fibrosis. J Clin Invest. 2018;128:45–53.
Wang H, Robinson JL, Kocabas P, Gustafsson J, Anton M, Cholley PE, Huang S, Gobom J, Svensson T, Uhlen M, Zetterberg H, Nielsen J. Genome-scale metabolic network reconstruction of model animals as a platform for translational research. Proc Natl Acad Sci U S A. 2021;118(30):e2102344118.
Ebrahim A, Lerman JA, Palsson BO, Hyduke DR. COBRApy: COnstraints-based reconstruction and analysis for python. BMC Syst Biol. 2013;7:74.
Maher P. The effects of stress and aging on glutathione metabolism. Ageing Res Rev. 2005;4:288–314.
You M, Arteel GE. Effect of ethanol on lipid metabolism. J Hepatol. 2019;70:237–48.
Softic S, Cohen DE, Kahn CR. Role of dietary fructose and hepatic de novo lipogenesis in fatty liver disease. Dig Dis Sci. 2016;61:1282–93.
Schreiter T, Gieseler RK, Vilchez-Vargas R, Jauregui R, Sowa JP, Klein-Scory S, Broering R, Croner RS, Treckmann JW, Link A, Canbay A. Transcriptome-wide analysis of human liver reveals age-related differences in the expression of select functional gene clusters and evidence for a PPP1R10-Governed ‘Aging Cascade’. Pharmaceutics. 2021;13(12):2009.
Karczewski KJ, Snyder MP. Integrative omics for health and disease. Nat Rev Genet. 2018;19:299–310.
Traag VA, Waltman L, van Eck NJ. From Louvain to Leiden: guaranteeing well-connected communities. Sci Rep. 2019;9:5233.
Lee S, Zhang C, Liu Z, Klevstig M, Mukhopadhyay B, Bergentall M, Cinar R, Stahlman M, Sikanic N, Park JK, Deshmukh S, Harzandi AM, Kuijpers T, Grotli M, Elsasser SJ, Piening BD, Snyder M, Smith U, Nielsen J, Backhed F, Kunos G, Uhlen M, Boren J, Mardinoglu A. Network analyses identify liver-specific targets for treating liver diseases. Mol Syst Biol. 2017;13:938.
Uhlen M, Fagerberg L, Hallstrom BM, Lindskog C, Oksvold P, Mardinoglu A, Sivertsson A, Kampf C, Sjostedt E, Asplund A, Olsson I, Edlund K, Lundberg E, Navani S, Szigyarto CA, Odeberg J, Djureinovic D, Takanen JO, Hober S, Alm T, Edqvist PH, Berling H, Tegel H, Mulder J, Rockberg J, Nilsson P, Schwenk JM, Hamsten M, von Feilitzen K, Forsberg M, Persson L, Johansson F, Zwahlen M, von Heijne G, Nielsen J, Ponten F. Proteomics. Tissue-based map of the human proteome. Science. 2015;347:1260419.
MacParland SA, Liu JC, Ma XZ, Innes BT, Bartczak AM, Gage BK, Manuel J, Khuu N, Echeverri J, Linares I, Gupta R, Cheng ML, Liu LY, Camat D, Chung SW, Seliga RK, Shao Z, Lee E, Ogawa S, Ogawa M, Wilson MD, Fish JE, Selzner M, Ghanekar A, Grant D, Greig P, Sapisochin G, Selzner N, Winegarden N, Adeyi O, Keller G, Bader GD, McGilvray ID. Single cell RNA sequencing of human liver reveals distinct intrahepatic macrophage populations. Nat Commun. 2018;9:4383.
Ungvari Z, Tarantini S, Donato AJ, Galvan V, Csiszar A. Mechanisms of vascular aging. Circ Res. 2018;123:849–67.
Dai DF, Rabinovitch PS, Ungvari Z. Mitochondria and cardiovascular aging. Circ Res. 2012;110:1109–24.
Balaban RS, Nemoto S, Finkel T. Mitochondria, oxidants, and aging. Cell. 2005;120:483–95.
Hunt NJ, Kang SWS, Lockwood GP, Le Couteur DG, Cogger VC. Hallmarks of aging in the liver. Comput Struct Biotechnol J. 2019;17:1151–61.
Ward W, Richardson A. Effect of age on liver protein synthesis and degradation. Hepatology. 1991;14:935–48.
Katzmann JL, Gouni-Berthold I, Laufs U. PCSK9 inhibition: insights from clinical trials and future prospects. Front Physiol. 2020;11:595819.
Spolitu S, Dai W, Zadroga JA, Ozcan L. Proprotein convertase subtilisin/kexin type 9 and lipid metabolism. Curr Opin Lipidol. 2019;30:186–91.
Lee JS, O'Connell EM, Pacher P, Lohoff FW. PCSK9 and the gut-liver-brain axis: a novel therapeutic target for immune regulation in alcohol use disorder. J Clin Med. 2021;10(8):1758.
Lee JS, Mukhopadhyay P, Matyas C, Trojnar E, Paloczi J, Yang YR, Blank BA, Savage C, Sorokin AV, Mehta NN, Vendruscolo JCM, Koob GF, Vendruscolo LF, Pacher P, Lohoff FW. PCSK9 inhibition as a novel therapeutic target for alcoholic liver disease. Sci Rep. 2019;9:17167.
Lohoff FW, Sorcher JL, Rosen AD, Mauro KL, Fanelli RR, Momenan R, Hodgkinson CA, Vendruscolo LF, Koob GF, Schwandt M, George DT, Jones IS, Holmes A, Zhou Z, Xu MJ, Gao B, Sun H, Phillips MJ, Muench C, Kaminsky ZA. Methylomic profiling and replication implicates deregulation of PCSK9 in alcohol use disorder. Mol Psychiatry. 2018;23:1900–10.
He Y, Rodrigues RM, Wang X, Seo W, Ma J, Hwang S, Fu Y, Trojnar E, Matyas C, Zhao S, Ren R, Feng D, Pacher P, Kunos G, Gao B. Neutrophil-to-hepatocyte communication via LDLR-dependent miR-223-enriched extracellular vesicle transfer ameliorates nonalcoholic steatohepatitis. J Clin Invest. 2021;131(3):e141513.
Schwartz GG, Steg PG, Szarek M, Bhatt DL, Bittner VA, Diaz R, Edelberg JM, Goodman SG, Hanotin C, Harrington RA, Jukema JW, Lecorps G, Mahaffey KW, Moryusef A, Pordy R, Quintero K, Roe MT, Sasiela WJ, Tamby JF, Tricoci P, White HD, Zeiher AM, Committees OO, Investigators,. Alirocumab and cardiovascular outcomes after acute coronary syndrome. N Engl J Med. 2018;379:2097–107.
Ray KK, Wright RS, Kallend D, Koenig W, Leiter LA, Raal FJ, Bisch JA, Richardson T, Jaros M, Wijngaard PLJ, Kastelein JJP, Orion & Investigators O. Two phase 3 trials of inclisiran in patients with elevated LDL cholesterol. N Engl J Med. 2020;382:1507–19.
Ballantyne CM, Banka P, Mendez G, Garcia R, Rosenstock J, Rodgers A, Mendizabal G, Mitchel Y, Catapano AL. Phase 2b randomized trial of the oral PCSK9 inhibitor MK-0616. J Am Coll Cardiol. 2023;81:1553–64.
Matyas C, Trojnar E, Zhao S, Arif M, Mukhopadhyay P, Kovacs A, Fabian A, Tokodi M, Bagyura Z, Merkely B, Kohidai L, Lajko E, Takacs A, He Y, Gao B, Paloczi J, Lohoff FW, Haskó G, Ding WX, Pacher P. PCSK9, a promising novel target for age-related cardiovascular dysfunction. J Am Coll Cardiol Basic Trans Science. 2023. https://doi.org/10.1016/j.jacbts.2023.06.005.
Pedregosa F, Varoquaux G, Gramfort A, Michel V, Thirion B, Grisel O, Blondel M, Prettenhofer P, Weiss R, Dubourg V, Vanderplas J, Passos A, Cournapeau D, Brucher M, Perrot M, Duchesnay E. Scikit-learn: Machine learning in python. J Mach Learn Res. 2011;12:2825–30.
Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol. 2014;15:550.
Varemo L, Nielsen J, Nookaew I. Enriching the gene set analysis of genome-wide data by incorporating directionality of gene expression and combining statistical hypotheses and methods. Nucleic Acids Res. 2013;41:4378–91.
Kuleshov MV, Jones MR, Rouillard AD, Fernandez NF, Duan Q, Wang Z, Koplev S, Jenkins SL, Jagodnik KM, Lachmann A, McDermott MG, Monteiro CD, Gundersen GW, Ma’ayan A. Enrichr: a comprehensive gene set enrichment analysis web server 2016 update. Nucleic Acids Res. 2016;44:W90-97.
Chen EY, Tan CM, Kou Y, Duan Q, Wang Z, Meirelles GV, Clark NR, Ma’ayan A. Enrichr: interactive and collaborative HTML5 gene list enrichment analysis tool. BMC Bioinformatics. 2013;14:128.
Mukhopadhyay P, Rajesh M, Cao Z, Horvath B, Park O, Wang H, Erdelyi K, Holovac E, Wang Y, Liaudet L, Hamdaoui N, Lafdil F, Hasko G, Szabo C, Boulares AH, Gao B, Pacher P. Poly (ADP-ribose) polymerase-1 is a key mediator of liver inflammation and fibrosis. Hepatology. 2014;59:1998–2009.
Bray NL, Pimentel H, Melsted P, Pachter L. Near-optimal probabilistic RNA-seq quantification. Nat Biotechnol. 2016;34:525–7.
Acknowledgements
This work utilized the computational resources of the NIH HPC Biowulf cluster (http://hpc.nih.gov).
Funding
The Genotype-Tissue Expression (GTEx) Project was supported by the Common Fund of the Office of the Director of the National Institutes of Health, and by NCI, NHGRI, NHLBI, NIDA, NIMH, and NINDS. The research was funded by Intramural Program of the Intramural Program of the National Institute on Alcohol Abuse and Alcoholism, National Institutes of Health to P.P.
Author information
Authors and Affiliations
Contributions
Conceptualization: M.A., C.M., P.P.; methodology: M.A., C.M., E.T., S.Z.; software: M.A.; formal analysis: M.A., C.M.; investigation: M.A, C.M., P.P.; validation: B.Y., P.M., J.P., B.P.L.; data curation: M.A.; writing—original draft: M.A, P.P.; writing—review and editing: M.A, C.M., P.M, B.Y., E.T., J.P., B.P.L., F.W.L, G.H., P.P; visualization: M.A.; supervision: P.P.; project administration: C.M, P.M.; funding acquisition: P.P.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
About this article
Cite this article
Arif, M., Matyas, C., Mukhopadhyay, P. et al. Data-driven transcriptomics analysis identifies PCSK9 as a novel key regulator in liver aging. GeroScience 45, 3059–3077 (2023). https://doi.org/10.1007/s11357-023-00928-w
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11357-023-00928-w