Abstract
Introduction
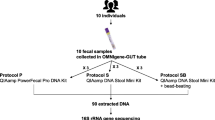
Fecal samples are highly complex and heterogeneous, containing materials at various stages of digestion. The heterogeneity and complexity of feces make stool metabolomics inherently challenging. The level of homogenization influences the outcome of the study, affecting the metabolite profiles and reproducibility; however, there is no consensus on how fecal samples should be prepared to overcome the topographical discrepancy and obtain data representative of the stool as a whole.
Objectives
Various combinations of homogenization conditions were compared to investigate the effects of bead size, addition of solvents and the differences between wet-frozen and lyophilized feces.
Methods
The homogenization parameters were systematically altered to evaluate the solvent usage, bead size, and whether lyophilization is required in homogenization. The metabolic coverage and reproducibility were compared among the different conditions.
Results
The current work revealed that a combination of mechanical and chemical lysis obtained by bead-beating with a mixture of big and small sizes of beads in an organic solvent is an effective way to homogenize fecal samples with adequate reproducibility and metabolic coverage. Lyophilization is required when bead-beating is not available.
Conclusions
A comprehensive and systematical evaluation of various fecal matter homogenization conditions provides a profound understanding for the effects of different homogenization methods. Our findings would be beneficial to assist with standardization of fecal sample homogenization protocol.



Similar content being viewed by others
References
Bliss, D. Z., Savik, K., Jung, H., Jensen, L., LeMoine, M., & Lowry, A. (1999). Comparison of subjective classification of stool consistency and stool water content. Journal of WOCN, 26(3), 137–141. https://doi.org/10.1016/S1071-5754(99)90031-1
Gangadoo, S., Rajapaksha Pathirannahalage, P., Cheeseman, S., Dang, Y. T. H., Elbourne, A., Cozzolino, D., Latham, K., Truong, V. K., & Chapman, J. (2021). The multiomics analyses of fecal matrix and its significance to coeliac disease gut profiling. International Journal of Molecular Sciences, 22(4), 1965. https://doi.org/10.3390/ijms22041965
Gao, X., Pujos-Guillot, E., & Sébédio, J.-L. (2010). Development of a quantitative metabolomic approach to study clinical human fecal water metabolome based on trimethylsilylation derivatization and GC/MS analysis. Analytical Chemistry, 82(15), 6447–6456. https://doi.org/10.1021/ac1006552
Gorzelak, M. A., Gill, S. K., Tasnim, N., Ahmadi-Vand, Z., Jay, M., & Gibson, D. L. (2015). Methods for improving human gut microbiome data by reducing variability through sample processing and storage of stool. PLoS ONE, 10(8), e0134802. https://doi.org/10.1371/journal.pone.0134802
Gratton, J., Phetcharaburanin, J., Mullish, B. H., Williams, H. R. T., Thursz, M., Nicholson, J. K., Holmes, E., Marchesi, J. R., & Li, J. V. (2016). Optimized sample handling strategy for metabolic profiling of human feces. Analytical Chemistry, 88(9), 4661–4668. https://doi.org/10.1021/acs.analchem.5b04159
Heaton, K. W., Radvan, J., Cripps, H., Mountford, R. A., Braddon, F. E., & Hughes, A. O. (1992). Defecation frequency and timing, and stool form in the general population: A prospective study. Gut, 33(6), 818–824. https://doi.org/10.1136/gut.33.6.818
Higgins Keppler, E. A., Jenkins, C. L., Davis, T. J., & Bean, H. D. (2018). Advances in the application of comprehensive two-dimensional gas chromatography in metabolomics. TrAC Trends in Analytical Chemistry, 109, 275–286. https://doi.org/10.1016/j.trac.2018.10.015
Karu, N., Deng, L., Slae, M., Guo, A. C., Sajed, T., Huynh, H., Wine, E., & Wishart, D. S. (2018). A review on human fecal metabolomics: Methods, applications and the human fecal metabolome database. Analytica Chimica Acta, 1030, 1–24. https://doi.org/10.1016/j.aca.2018.05.031
Lewis, S. J., & Heaton, K. W. (1997). Stool form scale as a useful guide to intestinal transit time. The Scandinavian Journal of Gastroenterolog, 32(9), 920–924. https://doi.org/10.3109/00365529709011203
Liang, Y., Dong, T., Chen, M., He, L., Wang, T., Liu, X., Chang, H., Mao, J.-H., Hang, B., Snijders, A. M., & **a, Y. (2020). Systematic analysis of impact of sampling regions and storage methods on fecal gut microbiome and metabolome profiles. msphere, 5(1), 10–1128. https://doi.org/10.1128/mSphere.00763-19
Lim, M. Y., Park, Y.-S., Kim, J.-H., & Nam, Y.-D. (2020). Evaluation of fecal DNA extraction protocols for human gut microbiome studies. BMC Microbiology, 20(1), 212. https://doi.org/10.1186/s12866-020-01894-5
Moosmang, S., Pitscheider, M., Sturm, S., Seger, C., Tilg, H., Halabalaki, M., & Stuppner, H. (2019). Metabolomic analysis—addressing NMR and LC-MS related problems in human feces sample preparation. Clinica Chimica Acta, 489, 169–176. https://doi.org/10.1016/j.cca.2017.10.029
Nam, S. L., de la Mata, A. P., Dias, R. P., & Harynuk, J. J. (2020). Towards standardization of data normalization strategies to improve urinary metabolomics studies by GC×GC-TOFMS. Metabolites, 10(9), 376. https://doi.org/10.3390/metabo10090376
O’Sullivan, V., Madrid-Gambin, F., Alegra, T., Gibbons, H., & Brennan, L. (2018). Impact of sample storage on the NMR fecal water metabolome. ACS Omega, 3(12), 16585–16590. https://doi.org/10.1021/acsomega.8b01761
Rose, C., Parker, A., Jefferson, B., & Cartmell, E. (2015). The characterization of feces and urine: A review of the literature to inform advanced treatment technology. Critical Reviews in Environmental Science and Technology, 45(17), 1827–1879. https://doi.org/10.1080/10643389.2014.1000761
Santiago, A., Panda, S., Mengels, G., Martinez, X., Azpiroz, F., Dore, J., Guarner, F., & Manichanh, C. (2014). Processing faecal samples: A step forward for standards in microbial community analysis. BMC Microbiology, 14(1), 112. https://doi.org/10.1186/1471-2180-14-112
Singh, R. K., Chang, H.-W., Yan, D., Lee, K. M., Ucmak, D., Wong, K., Abrouk, M., Farahnik, B., Nakamura, M., Zhu, T. H., Bhutani, T., & Liao, W. (2017). Influence of diet on the gut microbiome and implications for human health. Journal of Translational Medicine, 15(1), 73. https://doi.org/10.1186/s12967-017-1175-y
Smith, L., Villaret-Cazadamont, J., Claus, S. P., Canlet, C., Guillou, H., Cabaton, N. J., & Ellero-Simatos, S. (2020). Important considerations for sample collection in metabolomics studies with a special focus on applications to liver functions. Metabolites, 10(3), 104. https://doi.org/10.3390/metabo10030104
Vernocchi, P., Del Chierico, F., & Putignani, L. (2020). Gut microbiota metabolism and interaction with food components. International Journal of Molecular Sciences, 21(10), 3688. https://doi.org/10.3390/ijms21103688
Vijay, A., & Valdes, A. M. (2022). Role of the gut microbiome in chronic diseases: A narrative review. European Journal of Clinical Nutrition, 76(4), 489–501. https://doi.org/10.1038/s41430-021-00991-6
Wu, J., Wang, K., Wang, X., Pang, Y., & Jiang, C. (2021). The role of the gut microbiome and its metabolites in metabolic diseases. Protein & Cell, 12(5), 360–373. https://doi.org/10.1007/s13238-020-00814-7
Wu, W.-K., Chen, C.-C., Panyod, S., Chen, R.-A., Wu, M.-S., Sheen, L.-Y., & Chang, S.-C. (2019). Optimization of fecal sample processing for microbiome study—The journey from bathroom to bench. Journal of the Formosan Medical Association, 118(2), 545–555. https://doi.org/10.1016/j.jfma.2018.02.005
Yang, Y., Yin, Y., Chen, X., Chen, C., **a, Y., Qi, H., Baker, P. N., Zhang, H., & Han, T.-L. (2019). Evaluating different extraction solvents for GC-MS based metabolomic analysis of the fecal metabolome of adult and baby giant pandas. Science Reports, 9(1), 12017. https://doi.org/10.1038/s41598-019-48453-1
Zhang, B., Brock, M., Arana, C., Dende, C., Hooper, L., & Raj, P. (2020). Impact of bead-beating intensity on microbiome recovery in mouse and human stool: Optimization of DNA extraction. biorxiv. https://doi.org/10.1101/2020.06.15.151753
Funding
Authors would like to thank MITACS, DNA Genotek, Inc., The Natural Sciences and Engineering Research Council of Canada (NSERC) for support. The support of The Canada Foundation for Innovation (CFI), Genome Canada, and Genome Alberta to The Metabolomics Innovation Center (TMIC) is also acknowledged.
Author information
Authors and Affiliations
Contributions
All authors contributed to the study conception and design. Sample preparation and data collection were performed by KT; data processing and analysis were performed by KT and SLN. The first draft of the manuscript was written by KT and all authors commented on previous versions of the manuscript. All authors have read and agreed to the final manuscript.
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
This study was approved by the University of Alberta Research Ethics Board, under Approval number Pro00071285.
Informed consent
Informed consent was obtained from all individual participants included in this study.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary Information
Below is the link to the electronic supplementary material.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Tarazona Carrillo, K., Nam, S.L., de la Mata, A.P. et al. Optimization of fecal sample homogenization for untargeted metabolomics. Metabolomics 19, 74 (2023). https://doi.org/10.1007/s11306-023-02036-4
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11306-023-02036-4