Abstract
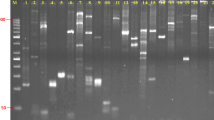
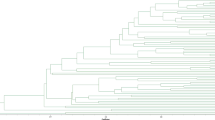
Oil camellia (Camellia oleifera Abel.) is a widely cultivated economically important plant. In China, more than 100 elite cultivars have been authorized for commercial plantations, representing the developmental base for the oil camellia industry. Since oil camellia seedlings are frequently traded and introduced to new places, there is an active demand to develop a rapid and reliable identification toolkit to protect and ensure the effective use of these resources. In this study, ten highly polymorphic simple sequence repeat (SSR) primer pairs were screened from the SSR primer database of oil camellia, and DNA fingerprints of 128 elite oil camellia cultivars were constructed with these primer pairs. By analyzing the fingerprinting profiles, all of these cultivars were found to have unique DNA fingerprints, except ‘Changlin 40’ and ‘Fuyang 40’, which shared a complete similarity across all of the fingerprints generated by these ten SSR primer pairs, indicating that they were either synonyms or vegetative mutants. Synonyms are a major source of confusion in protecting and managing new varieties of plants. Although the synonymy of these two cultivars needs to be further confirmed, our data indicated a high possibility of synonyms among the authorized oil camellia cultivars. This study provides a desirable genetic toolkit for the identification and management of the elite oil camellia cultivars, and also for the intellectual property right protection of breeders.



Similar content being viewed by others
References
Botstein D, White RL, Skolnick M, Davis RW (1980) Construction of genetic linkage map in man using restriction fragment length polymorphisms. Am J Hum Genet 32:314–331
Chen YZ (2006) Tea oil camellia. In: Hu FM, Tan XF, Liu HM (eds) Culture and utilization of Chinese non-wood product forest trees. China forestry publishing house, Bei**g, pp 370–383
Chen YZ, Zhang ZY, Tan XF (2005) Identification of oil tea (Camellia oleifera) superior clones by RAPD molecular marker. J Central South Forestry University 25:40–45. doi:10.14067/j.cnki.1673 -923x.2005.04.009
Chen H, He M, Huang Q, Liu D, Huang M (2007) Sasanquasaponin protects rat cardiomycytes against oxidatice stress induced by anoxia-reoxygenation injury. Eur J Pharmacol 575:21–27. doi:10.1016/j.ejphar.2007.07.043
Chen YF, Yang CH, Chang MS et al (2010) Foam properties and detergent abilities of the saponins from Camellia oleifera. Int J Mol Sci 11:4417–4425. doi:10.3390/ijms11114417
Felsenstein J (1989) PHYLIP-phylogeny inference package. Cladistics 5:164–166. doi:10.1111/j.1096-0031.1989.tb00562.x
He L, Guo YZ, Huai YZ et al (2011) Research progress on the health function of tea oil. J Med Plant Res 5:485–489
Huang SF, Zhao ZF, Wu RJ, Li JP, Hsu PS (1984) Karyotype analysis in Camellia chekiangoleosa Hu. Guihaia 4:285–288
Huang Q, Shao L, He M, Chen H, Liu D, Luo Y, Dai Y (2005) Inhibitory effects of sasanquasaponin on over-expression of ICAM-1 and on enhancement of capillary permeability induced by burns in rats. Burns 31:637–642. doi:10.1016/j.burns.2005.02.006
Huang YF, Chen X, Zhuang XY et al (2006) Analysis of genetic diversity in Camellia oleifera germplasms. Scientia silvae sinicae 42:38–43. doi:10.11707/j.1001-7488.20060407
Huang Q, He M, Chen H, Shao L, Liu D, Luo Y, Dai Y (2007) Protective effects of sasanquasaponin on injury of endothelial cells induced by anoxia and reoxygenation in vitro. Basic Clin Pharmacol Toxicol 101:301–308. doi:10.1111/j.1742-7843.2007.00119.x
Junichi T, Fumiya T, Noriko H, Satoshi Y (2006) Estimation of the genome size of tea (Camellia sinensis) camellia (C. japonica) and their interspecific hybrids by flow cytometry. Chagyo Kenkyu Hokoku 101:1–7. doi:10.5979/cha.2006.1
Kaur S, Panesar PS, Bera MB et al (2015) Simple sequence repeat markers in genetic divergence and marker-assisted selection of rice cultivars: a review. Crit Rev Food Sci Nutr 55:41–49. doi:10.1080/10408398.2011.646363
Keim P, Beavis W, Schupp J, Freestone R (1992) Evaluation of soybean RFLP marker diversity in adapted germplasm. Theor Appl Genet 85:205–212. doi:10.1007/BF00222861
Korir NK, Han J, Shangguan L et al (2013) Plant variety and cultivar identification: advances and prospects. Crit Rev Biotechnol 33:111–125. doi:10.3109/07388551.2012.675314
Kunihisa M, Ueda H, Fukino N, Matsumoto S (2009) DNA markers for identification of strawberry (Fragaria × ananassa Duch.) cultivars based on probability theory. J Jpn Soc Hortic Sci 78:211–217
Li GT (2001) New advance in karyotype studies of genus Camellia. Chin Wild Plant Resour 20:9--14
Lin P, Yao XH, Wang KL, Zheng TT, Teng JH (2010) Idengtification and genetic analysis of camellia oleifera changlin series superior clones by SRAP molecular marker. J Agric Biotechnol 18:272–279. doi:10.3969/j.issn.1674-7968.2010.02.011
Long Z, Wang D (2008) Chemical constituents of olive oil and from Camellia oleifera seed oil. J Chinese cereals and oils Assoc 23:121–123
Mondal TK (2011) Camellia. In: Kole C (ed) Wild crop relatives: genomic and breeding resources. Springer, Berlin Heidelberg, New York, pp 15–39
Morell MK, Peakall R, Appels R et al (1995) DNA profiling techniques for plant variety identification. Aust J Exp Agric 35:807–819. doi:10.1071/EA9950807
Mukhopadhyay M, Mondal TK (2014) The physio-chemical responses of Camellia plants to abiotic stresses. J Plant Sci Res 1:105
Oliveira MB, Vieira ESN, Schuster I (2010) Construction of a molecular database for soybean cultivar identification in Brazil. Genet Mol Res 9:705–720. doi:10.4238/vol9-2gmr706
Pan ZE, Sun J, Wang X, Jia Y, Zhou Z, Pang B, Du X (2008) Screening of SSR core primers with polymorphism on a cotton panel. Biodivers Sci 16:555–561. doi:10.3724/SP.J.1003.2008.08132
Que SQ, Zhang F (2008) DNA Extraction from Camellia oleifera and the establishment of the optimum reaction system of RAPD analysis. J Anhui agricultural Sci 36:902. doi:10.13989/j.cnki.0517-6611.2008.03.125
Selkoe KA, Toonen RJ (2006) Microsatellites for ecologists: a practical guide to using and evaluating microsatellite markers. Ecol Lett 9:615–629. doi:10.1111/j.1461-0248.2006.00889.x
Shi J, Dai X, Chen Y et al (2013) Discovery and experimental analysis of microsatellites in an oil woody plant Camellia chekiangoleosa. Plant Syst Evol 299:1387–1393. doi:10.1007/s00606-013-0814-x
Sun PG, ** RC, Li JC, Ouyang KX, Zhong HY, Chen XY (2014) Genetic analyses and molecular identification of 25 superior clones of Camellia oleifera. J South China Agric Univ 35:83–88. doi:10.7671/j.issn.1001-411X.2014.06.016
Tamura K, Stecher G, Peterson D, Filipski A, Kumar S (2013) MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol 30:2725–2729. doi:10.1093/molbev/mst197
The State Forestry Administration of China. 2009. Bei**g: National camellia oleifera industry development plan (2009–2020). National Plan for the Development of Camellia oleifera oil (2009–2020) http://www.gnycw.com/Law/ShowInfo.asp? InfoID = 523.
UPOV (International Union for the Protection of New Varieties of Plants) (2010) Guidelines for DNA profiling: molecular marker selection and database construction (“BMT guidelines”). UPOV, Geneva, Switzerland
Varshney RK, Graner A, Sorrells ME (2005) Genic microsatellite markers in plants: features and applications. Trends Biotechnol 23:48–55. doi:10.1016/j.tibtech.2004.11.005
Wang X, Chen Y, Wu L (2008) Oil content and fatty acid composition of Camellia oleifera seed. J Central South Univ Forestry Technol 28:11–17. doi:10.14067/j.cnki.1673-923x.2008.03.023
Wang L, Bai J, Wang Y, Wang Y, Zhao W, Su L (2010) Selection of SSR core primers for Maize hybrid purity identification in Shanxi Province. J Shanxi Agric Sci. doi:10.3969/j.issn.1002-2481.2010.01.05
Wang FG, Tian HL, Zhao JR, et al (2011) Development and characterization of a core set of SSR markers for fingerprinting analysis of Chinese maize varieties. Maydica, 56.
Wei L, Zhao C, Li J et al (2012) Screening of core simple sequence repeat primer pairs and establishment of a multiplex polymerase chain reaction system for Brassica genomes A and C. Plant Breed 131:457–459. doi:10.1111/j.1439-0523.2012.01955.x
Wen Q, Lei X, Ye J et al (2008) Identification of Camellia oleifera superior clones by ISSR molecular markers. J Central South Univ Forestry Technol 1:008. doi:10.14067/j.cnki.1673-923x.2008.01.024
Wen Q, Xu L, Gu Y et al (2012) Development of polymorphic microsatellite markers in Camellia chekiangoleosa (Theaceae) using 454-ESTs. Am J Bot 99:e203–e205. doi:10.3732/ajb.1100486
Wünsch A, Hormaza JI (2002) Cultivar identification and genetic fingerprinting of temperate fruit tree species using DNA markers. Euphytica 125:59–67. doi:10.1023/A:1015723805293
**a EH, Jiang JJ, Huang H et al (2014) Transcriptome analysis of the oil-rich tea plant, camellia oleifera, reveals candidate genes related to lipid metabolism. PLoS One 9:e104150. doi:10.1371/journal.pone.0104150
**ng C, Liu X, ** Q et al (2012) Moisture sorption thermodynamics of camellia oleifera. Food biophysics 7:163–172. doi:10.1007/s11483-012-9254-3
Yu X, Yu F, Liu J, Chen J (2013) Identification and genetic diversity analysis of Camellia oleifera varieties using ISSR marker. J Nan**g Forestry Univ(Natural Sci Ed) 37:61–66
Zhang ZJ (2002) Tissue culture, molecular identification by RAPD markers and construction of cDNA library in tea oil camellia. Central South Forestry University, Dissertation
Zhang G, Zhong W, Wuyun T et al (2007) Identification of oil tea Camellia oleifera superior clones by ISSR molecular marker. For Res 20:278–283
Zhong HY, **e BX, Wan CN (2001) Current situation and prospect on utilization of Camellia oleifera in China. China Forestry Sci Technol 15:6–8
Zhuang RL, Dong RX (1984) The preliminary study on size and variation of pollens and on chromosome numbers of Camellia spp. For Sci Technol 3:15–17. doi:10.13456/j.cnki.lykt.1984.03.007
Acknowledgments
We thank Prof. Shuxian Li and Jiseng Shi at the Southern Forest Tree Seeds Inspection Center of China for their help in collecting the elite oil camellia cultivars. This work was funded by the 948 Project (2014-4-13), the Natural Science Foundation of China and Jiangsu Province (31400564, 31270711, and BK20130968). It was also supported by Jiangsu Planned Projects for Postdoctoral Research Funds (1402011B), the Innovative Research Team Program in Jiangsu Province and the Educational Department of China, and the Priority Academic Program Development (PAPD) Program at Nan**g Forestry University.
Author information
Authors and Affiliations
Corresponding author
Additional information
Communicated by M. Wirthensohn
Yingnan Chen and **aogang Dai contributed equally to this work.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Table S1
Detailed information of the examined oil camellia cultivars.(XLS 21 kb)
Table S2
A summary of the allele sizes amplified in each cultivar by each of the SSR primers.(XLS 32 kb)
Table S3
Genotypes detected by each SSR primer and the included cultivars with the same genotype at each SSR locus.(XLS 42 kb)
Table S4
The frequency of each cultivar’s genotype at each of the SSR loci.(XLS 27 kb)
Rights and permissions
About this article
Cite this article
Chen, Y., Dai, X., Hou, J. et al. DNA fingerprinting of oil camellia cultivars with SSR markers. Tree Genetics & Genomes 12, 7 (2016). https://doi.org/10.1007/s11295-015-0966-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11295-015-0966-7