Abstract
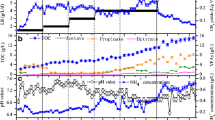
In this study, rumen content was used to obtain three enrichments of anaerobic fungi and methanogens (F + M enrichment), bacteria and methanogens (B + M enrichment), and whole rumen content (WRC enrichment), to evaluate their respective ability to degrade lignocellulose and produce methane. Among the treatments, F + M enrichment elicited the strongest lignocellulose degradation and methane production ability with both rice straw and wheat straw as substrates. Quantitative real-time PCR analysis and diversity analyses of methanogens in the three enrichment treatments demonstrated that F + M had larger number of 16S rRNA gene copies of methanogens and higher relative abundance of Methanobrevibacter, the predominant methanogen found in all enrichments. Caecomyces was the main anaerobic fungal genus for co-culturing to provide substrates for methanogens in this enrichment. Importantly, the F + M enrichment was stable and could be maintained with transfers supplied every 3 days, confirming its potential utility in anaerobic digestion for lignocellulose degradation and methane production.



Similar content being viewed by others
References
AOAC (1990) Official methods of analysis, 15th edn. AOAC International, Arlington
Akin DE, Lyon CE, Windham WR, Rigsby LL (1989) Physical degradation of lignified stem tissues by ruminal fungi. Appl Environ Microbiol 55(3):611–616
Barr DJS, Kudo H, Jakober KD, Cheng KJ (1989) Morphology and development of rumen fungi: Neocallimastix sp., Piromyces communis, and Orpinomyces bovis gen. nov., sp. nov. Can J Bot 67:2815–2824
Breton A, Bernalier A, Dusser M, Fonty G, Gaillard-Martinie B, Guillot J (1990) Anaeromyces mucronatus nov. gen., nov. sp. A new strictly anaerobic rumen fungus with polycentric thallus. FEMS Microbiol Lett 70:177–182
Callaghan TM, Podmirseg SM, Hohlweck D, Edwards JE, Puniya AK, Dagar SS, Griffith GW (2015) Buwchfawromyces eastonii gen nov, sp. nov: a new anaerobic fungus (Neocallimastigomycota) isolated from buffalo faeces. MycoKeys 9:11
Caporaso JG, Kuczynski J, Stombaugh J, Bittinger K, Bushman FD, Costello EK, Fierer N, Pena AG, Goodrich JK, Gordon JI, Huttley GA, Kelley ST, Knights D, Koenig JE, Ley RE, Lozupone CA, McDonald D, Muegge BD, Pirrung M, Reeder J, Sevinsky JR, Turnbaugh PJ, Walters WA, Widmann J, Yatsunenko T, Zaneveld J, Knight R (2010) QIIME allows analysis of high-throughput community sequencing data. Nat Methods 7:335–336
Cersosimo LM, Lachance H, St-Pierre B, van Hoven W, Wright AD (2015) Examination of the rumen bacteria and methanogenic archaea of wild impalas (Aepyceros melampus melampus) from Pongola, South Africa. Microb Ecol 69:577–585
Cheng YF, Edwards JE, Allison GG, Zhu WY, Theodorou MK (2009) Diversity and activity of enriched ruminal cultures of anaerobic fungi and methanogens grown together on lignocellulose in consecutive batch culture. Bioresour Technol 100:4821–4828
Cheng Y, Shi Q, Sun R, Liang D, Li Y, Li Y, ** W, Zhu W (2018) The biotechnological potential of anaerobic fungi on fiber degradation and methane production. World J Microbiol Biotechnol 34:155
Couger MB, Youssef NH, Struchtemeyer CG, Liggenstoffer AS, Elshahed MS (2015) Transcriptomic analysis of lignocellulosic biomass degradation by the anaerobic fungal isolate Orpinomyces sp. strain C1A. Biotechnol Biofuels 8:208
Dagar SS, Kumar S, Griffith GW, Edwards JE, Callaghan TM, Singh R, Nagpal AK, Puniya AK (2015) A new anaerobic fungus (Oontomyces anksri gen. nov., sp. nov.) from the digestive tract of the Indian camel (Camelus dromedarius). Fungal Biol 119:731–737
Dridi B, Fardeau ML, Ollivier B, Raoult D, Drancourt M (2012) Methanomassiliicoccus luminyensis gen nov, sp nov, a methanogenic archaeon isolated from human faeces. Int J Syst Evol Microbiol 62(Pt 8):1902–1907
Ellabban O, Abu-Rub H, Blaabjerg F (2014) Renewable energy resources: current status, future prospects and their enabling technology. Renew Sust Energ Revi 39:748–764
Gold JJ, Heath IB, Bauchop T (1988) Ultrastructural description of a new chytrid genus of caecum anaerobe, Caecomyces equi gen. nov., sp. nov., assigned to the Neocallimasticaceae. Biosystems 21:403–415
Hanafy RA, Elshahed MS, Liggenstoffer AS, Griffith GW, Youssef NH (2017) Pecoramyces ruminantium, gen. nov., sp. nov., an anaerobic gut fungus from the feces of cattle and sheep. Mycologia 109:231–243
Hanafy RA, Elshahed MS, Youssef NH (2018) Feramyces austinii, gen. nov., sp. nov., an anaerobic gut fungus from rumen and fecal samples of wild Barbary sheep and fallow deer. Mycologia 110:513–525
He X, Wu Y, Cai M, Mu C, Luo W, Cheng Y, Zhu W (2015) The effect of increased atmospheric temperature and CO2 concentration during crop growth on the chemical composition and in vitro rumen fermentation characteristics of wheat straw. J Anim Sci Biotechnol 6:46
Heath IB, Bauchop T, Skipp RA (1983) Assignment of the rumen anaerobe Neocallimastix frontalis to the Spizellomycetales (Chytridiomycetes) on the basis of its polyflagellate zoospore ultrastructure. Can J Bot 61:295–307
Henske JK, Gilmore SP, Knop D, Cunningham FJ, Sexton JA, Smallwood CR, Shutthanandan V, Evans JE, Theodorou MK, O’Malley MA (2017) Transcriptomic characterization of Caecomyces churrovis: a novel, non-rhizoid-forming lignocellulolytic anaerobic fungus. Biotechnol Biofuels 10:305
Huang J, Li Y (2018) Rumen methanogen and protozoal communities of Tibetan sheep and Gansu Alpine Finewool sheep grazing on the Qinghai-Tibetan Plateau, China. BMC Microbiol 18:212
Iino T, Tamaki H, Tamazawa S, Ueno Y, Ohkuma M, Suzuki K, Igarashi Y, Haruta S (2013) Candidatus Methanogranum caenicola: a novel methanogen from the anaerobic digested sludge, and proposal of Methanomassiliicoccaceae fam. nov. and Methanomassiliicoccales ord. nov., for a methanogenic lineage of the class Thermoplasmata. Microb Environ 28:244–250
** W, Cheng YF, Mao SY, Zhu WY (2011) Isolation of natural cultures of anaerobic fungi and indigenously associated methanogens from herbivores and their bioconversion of lignocellulosic materials to methane. Bioresour Technol 102:7925–7931
Joshi A, Lanjekar VB, Dhakephalkar PK, Callaghan TM, Griffith GW, Dagar SS (2018) Liebetanzomyces polymorphus gen. et sp. nov., a new anaerobic fungus (Neocallimastigomycota) isolated from the rumen of a goat. MycoKeys 40:89–110
Joblin KN, Campbell GP, Richardson AJ et al (1989) Fermentation of barley straw by anaerobic rumen bacteria and fungi in axenic culture and in co-culture with methanogens. Lett Appl Microbiol 9:195–197
Kittelmann S, Seedorf H, Walters WA, Clemente JC, Knight R, Gordon JI, Janssen PH (2013) Simultaneous amplicon sequencing to explore co-occurrence patterns of bacterial, archaeal and eukaryotic microorganisms in rumen microbial communities. PLoS ONE 8:e47879
Kumar P, Barrett DM, Delwiche MJ, Stroeve P (2009) Methods for pretreatment of lignocellulosic biomass for efficient hydrolysis and biofuel production. Ind Eng Chem Res 48:3713–3729
Lazuka A, Auer L, O’Donohue M, Hernandez-Raquet G (2018) Anaerobic lignocellulolytic microbial consortium derived from termite gut: enrichment, lignocellulose degradation and community dynamics. Biotechnol Biofuels 11:284
Li Y, ** W, Cheng Y, Zhu Y (2016) Effect of the associated methanogen Methanobrevibacter thaueri on the dynamic profile of end and intermediate metabolites of anaerobic fungus Piromyces sp. F1. Curr Microbiol 73:434–441
Li Y, ** W, Mu C, Cheng Y, Zhu W (2017) Indigenously associated methanogens intensified the metabolism in hydrogenosomes of anaerobic fungi with xylose as substrate. J Basic Microbiol 57:933–940
Lee S, Ha J, Cheng KJ (2000) Relative contributions of bacteria, protozoa, and fungi to in vitro degradation of orchard grass cell walls and their interactions. Appl Environ Microbiol 66(9):3807–3813
Miller TL, Lin C (2002) Description of Methanobervibacter gottschalkii sp. nov., Methanobrevibacter thaueri sp. nov., Methanobrevibacter woesei sp. nov. and Methanobrevibacter wolinii sp. nov. Int J Syst Evol Microbiol 52(Pt3):819–822
Nilsson RH, Larsson KH, Taylor AFS, Bengtsson-Palme J, Jeppesen TS, Schigel D, Kennedy P, Picard K, Glockner FO, Tedersoo L, Saar I, Koljalg U, Abarenkov K (2018) The UNITE database for molecular identification of fungi: handling dark taxa and parallel taxonomic classifications. Nucleic Acids Res. https://doi.org/10.1093/nar/gky1022
Ozkose E, Thomas BJ, Davies DR, Griffith GW, Theodorou MK (2001) Cyllamyces aberensis gen. nov. sp. nov., a new anaerobic gut fungus with branched sporangiophores isolated from cattle. Can J Bot 79:666–673
Park JG, Lee B, Park HR, Jun HB (2019) Long-term evaluation of methane production in a bio-electrochemical anaerobic digestion reactor according to the organic loading rate. Biores Technol 273:478–486
Paul SS, Bu D, Xu J, Hyde KD, Yu Z (2018) A phylogenetic census of global diversity of gut anaerobic fungi and a new taxonomic framework. Fungal Divers 89:1–14
Rea S, Bowman JP, Popovski S, Pimm C, Wright AD (2007) Methanobrevibacter millerae sp. nov. and Methanobrevibacter olleyae sp. nov., methanogens from the ovine and bovine rumen that can utilize formate for growth. Int J Syst Evol Microbiol 57(Pt 3):450–456
Seedorf H, Kittelmann S, Henderson G, Janssen PH (2014) RIM-DB: a taxonomic framework for community structure analysis of methanogenic archaea from the rumen and other intestinal environments. Peer J 2:e494
Solomon KV, Haitjema CH, Henske JK, Gilmore SP, Borges-Rivera D, Lipzen A, Brewer HM, Purvine SO, Wright AT, Theodorou MK, Grigoriev IV, Regev A, Thompson DA, O’Malley MA (2016) Early-branching gut fungi possess a large, comprehensive array of biomass-degrading enzymes. Science 351:1192–1195
Stopnisek N, Zuhlke D, Carlier A, Barberan A, Fierer N, Becher D, Riedel K, Eberl L, Weisskopf L (2016) Molecular mechanisms underlying the close association between soil Burkholderia and fungi. ISME J 10:253–264
Tapio I, Fischer D, Blasco L, Tapio M, Wallace RJ, Bayat AR, Ventto L, Kahala M, Negussie E, Shingfield KJ, Vilkki J (2017) Taxon abundance, diversity, co-occurrence and network analysis of the ruminal microbiota in response to dietary changes in dairy cows. PLoS ONE 12:e0180260
Theodorou MK, Davies DR, Nielsen BB et al (1995) Determination of growth of anaerobic fungi on soluble and cellulosic substrates using a pressure transducer. Microbiology 141(3):671–678
Van Soest PJ, Robertson JB, Lewis BA (1991) Methods for dietary fiber, neutral detergent fiber, and nonstarch polysaccharides in relation to animal nutrition. J Dairy Sci 74:3583–3597
Weatherburn M (1967) Phenol-hypochlorite reaction for determination of ammonia. Anal Chem 39:971–974
Weimer PJ, Russell JB, Muck RE (2009) Lessons from the cow: what the ruminant animal can teach us about consolidated bioprocessing of cellulosic biomass. Biores Technol 100:5323–5331
Yu Y, Lee C, Kim J, Hwang S (2005) Group-specific primer and probe sets to detect methanogenic communities using quantitative real-time polymerase chain reaction. Biotechnol Bioeng 89:670–679
Zhang G, Lu F, Zhao H, Yang G, Wang X, Ouyang Z (2017) Residue usage and farmers' recognition and attitude toward residue retention in China's croplands. J Agro-Environ Sci 36:981–988 (in Chinese)
Zhao L, Meng Q, Li Y, Wu H, Huo Y, Zhang X, Zhou Z (2018) Nitrate decreases ruminal methane production with slight changes to ruminal methanogen composition of nitrate-adapted steers. BMC Microbiol 18:21
Acknowledgements
This study was funded by the Natural Science Foundation of China (Grant Number 31772627), the “Belt and Road” Cooperation Program of Jiangsu Province (Grant Number BZ2018055) and the Fundamental Research Funds for the Central Universities (Grant Number KYDK201701).
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflict of interest
The authors declare that they have no conflict of interest.
Ethical approval
All applicable international, national, and/or institutional guidelines for the care and use of animals were followed.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Electronic supplementary material
Below is the link to the electronic supplementary material.
Rights and permissions
About this article
Cite this article
Ma, Y., Li, Y., Li, Y. et al. The enrichment of anaerobic fungi and methanogens showed higher lignocellulose degrading and methane producing ability than that of bacteria and methanogens. World J Microbiol Biotechnol 36, 125 (2020). https://doi.org/10.1007/s11274-020-02894-3
Received:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11274-020-02894-3