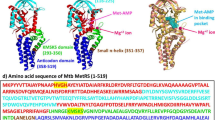
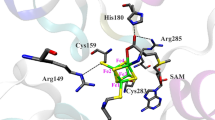
Abstract
Aminoacyl-tRNA synthetases (aaRS) are key enzymes in protein biosynthesis that employ various editing mechanisms to ensure faithful translation of genetic information. Seryl-tRNA synthetase (SerRS) and isoleucine-tRNA synthetase (IleRS) utilize pre-transfer editing mechanisms to prevent the incorporation of the undesired misactivated amino acids. In this study, molecular dynamics (MD) and quantum mechanics/QM (QM/semiempirical) were used to provide atomistic details regarding the pre-transfer editing against the non-cognates homocysteine (Hcy) by IleRS as well as cysteine (Cys) and threonine (Thr) by SerRS. Notably, in both enzymes, the mechanism is found to follow a stepwise self-cyclization substrate-assisted mechanism. Initially, dihedral scan around the substrate’s Cβ__Cγ bond takes place followed by a concerted step of R-S(O)H deprotonation concomitant with a nucleophilic attack. The rate limiting step to edit against Hcy by IleRS is the dihedral scan step and require 24.3 kcal/mol to proceed. Meanwhile, the rate-limiting step for editing against both Cys and Thr by SerRS is found to be the second step with Gibbs energy barriers of 20.4 and 26.6 kcal/mol, respectively. Interestingly, following the same pre-transfer editing pathway against the cognate Ser by SerRS results in a significantly higher energy barrier of 31.4 kcal/mol and energetically less favoured product complex lies at 17.7 kcal/mol. This finding clearly indicates an enzymatically infeasible process against Ser-AMP from kinetic and thermodynamic perspectives. The provided mechanistic insights should provide novel foundation required for the development of effective therapeutic agents for the treatment of the many disease states associated with these enzymes.








Similar content being viewed by others
Availability of data and materials
Coordinates for the optimized structures have been provided with the manuscript and more date can be provided once requested.
Abbreviations
- aaRS:
-
Aminoacyl-tRNA Synthetases
- SerRS:
-
Seryl-tRNA synthetases
- IleRS:
-
Isoleucine-tRNA synthetase
- Ser:
-
Serine
- Thr:
-
Threonine
- Hcy:
-
Homocysteine
- RC:
-
Reactive complex
- TS:
-
Transition state
- IC:
-
Intermediate complex
- PC:
-
Product complex
- Op:
-
Non-bridged phosphate oxygen
- H-bond:
-
Hydrogen bond
- ATP:
-
Adenosine triphosphate
- AMP:
-
Adenosine monophosphate
References
Moghal A, Mohler K, Ibba M et al (2014) Mistranslation of the genetic code. FEBS Lett 588:4305–4310
Kim S, You S, Hwang D et al (2011) Aminoacyl-tRNA synthetases and tumorigenesis: more than housekee**. Nat Rev Cancer 11:708–718
Yao P, Fox PL (2013) Aminoacyl-tRNA synthetases in medicine and disease. EMBO Mol Med 5:332–343
Nie A, Sun B, Fu Z, Yu D et al (2019) Roles of aminoacyl-tRNA synthetases in immune regulation and immune diseases. Cell Death Dis 10
Shim JA, Jo Y, Hwang H, Lee SE, Ha D, Lee JH, Kim J, Song P, Lee D, Hong C et al (2022) Defects in aminoacyl-tRNA synthetase cause partial B and T cell immunodeficiency. Cell Mol Life Sci 79
Eriani G, Delarue M, Poch O, Gangloff J, Moras D (1990) Partition of transfer-RNA synthetases into 2 classes based on mutually exclusive sets of sequence motifs. Nature 347:203–206
Perona JJ, Gruic-Sovulj I (2014) Synthetic and editing mechanisms of aminoacyl-tRNA synthetases. In Aminoacyl-tRNA synthetases in biology and medicine, Kim S Ed. 344:1–41
Banik SD, Nandi N (2012) Mechanism of the activation step of the aminoacylation reaction: a significant difference between class I and class II synthetases. J Biomol Struct Dynam 30(6):701–715
Yadavalli SS, Ibba M (2012) In Advances in protein chemistry and structural biology, Vol 86: fidelity and quality control in gene expression, Marintchev A Ed. 86:1–43
Schimmel P (2011) Mistranslation and its control by tRNA synthetases. Philoso Trans R Soc B Biol Sci 366:2965–2971
Hussain T, Kamarthapu V, Kruparani SP, Deshmukh MV, Sankaranarayanan R et al (2010) Mechanistic insights into cognate substrate discrimination during proofreading in translation. Proc Natl Acad Sci USA 107:22117–22121
Moras D (2010) Proofreading in translation: dynamics of the double-sieve model. Proc Natl Acad Sci USA 107:21949–21950
Cvetesic N, Dulic M, Bilus M, Sostaric N, Lenhard B, Gruic-Sovulj I et al (2016) Naturally Occurring isoleucyl-tRNA synthetase without tRNA-dependent pre-transfer editing. J Biol Chem 291:8618–8631
Hati S, Ziervogel B, SternJohn J, Wong FC, Nagan MC, Rosen AE, Siliciano PG, Chihade JW, Musier-Forsyth K et al (2006) Pre-transfer editing by class II prolyl-tRNA synthetase - role of aminoacylation active site in “selective release” of noncognate amino acids. J Biol Chem 38:27862–27872
Cvetesic N, Bilus M, Gruic-Sovulj I et al (2015) The tRNA A76 hydroxyl groups control partitioning of the tRNA-dependent pre- and post-transfer editing pathways in class I tRNA synthetase. J Biol Chem 290:13981–13991
Dulic M, Perona JJ, Gruic-Sovulj I et al (2014) Determinants for tRNA-dependent pretransfer editing in the synthetic site of isoleucyl-tRNA synthetase. Biochemistry 53:6189–6198
Dulic M, Cvetesic N, Perona JJ, Gruic-Sovulj I et al (2010) Partitioning of tRNA-dependent editing between pre- and post-transfer pathways in class I aminoacyl-tRNA synthetases. J Biol Chem 285(31):23799–23809
Jakubowski H, Goldman E (1992) Editing of errors in selection of amino-acids for protein-synthesis. Microbiol Rev 56:412–429
Fukunaga R, Yokoyama S (2006) Structural basis for substrate recognition by the editing domain of isoleucyl-tRNA synthetase. J Mol Biol 359:901–912
Nomanbhoy TK, Hendrickson TL, Schimmel P et al (1999) Transfer RNA-dependent translocation of misactivated amino acids to prevent errors in protein synthesis. Mol Cell 4:519–528
Jakubowski H (2001) Translational accuracy of aminoacyl-tRNA synthetases: implications for atherosclerosis. J Nut 131:2983S-2987S
Hartlein M, Cusack S (1995) Structure, function and evolution of seryl-transfer-rna synthetases - implications for the evolution of aminoacyl-transfer-RNA synthetases and the genetic-code. J Mol Evol 40:519–530
Gruic-Sovulj I, Rokov-Plavec J, Weygand-Durasevic I et al (2007) Hydrolysis of non-cognate amino acyl-adenylates by a class II aminoacyl-tRNA synthetase lacking an editing domain. FEBS Lett 581:5110–5114
Bilokapic S, Maier T, Ahel D, Gruic-Sovulj I, Soll D, Weygand-Durasevic I, Ban N et al (2006) Structure of the unusual seryl-tRNA synthetase reveals a distinct zinc-dependent mode of substrate recognition. EMBO J 25:2498–2509
Xu XM, Carlson BA, Mix H, Zhang Y, Saira K, Glass RS, Berry MJ, Gladyshev VN, Hatfield DL et al (2007) Biosynthesis of selenocysteine on its tRNA in eukaryotes. Faseb J 21:A113–A113
Huang WJ, Bushnell EAC, Francklyn CS, Gauld JW et al (2011) The alpha-amino group of the threonine substrate as the general base during tRNA aminoacylation: a new version of substrate-assisted catalysis predicted by hybrid DFT. J Phys Chem A 115:13050–13060
Liu HN, Gauld JW (2008) Substrate-assisted catalysis in the aminoacyl transfer mechanism of histidyl-tRNA synthetase: a density functional theory study. J Phys Chem B 112:16874–16882
Aboelnga MM, Gauld JW (2017) Roles of the active site Zn(II) and residues in substrate discrimination by threonyl-tRNA synthetase: an MD and QM/MM investigation. J Phys Chem B 121:6163–6174
Aboelnga MM, Gauld JW, Hayward JJ et al (2017) A water-mediated and substrate-assisted aminoacylation mechanism in the discriminating aminoacyl-tRNA synthetase GlnRS and non-discriminating GluRS. Phys Chem Chem Phys 19:25598–25609
Aboelnga MM, Hayward JJ, Gauld JW et al (2018) Unraveling the critical role played by Ado762′OH in the post-transfer editing by archaeal threonyl-tRNA synthetase. J Phys Chem B 122:1092–1101
Fortowsky GB, Simard DJ, Aboelnga MM, Gauld JW et al (2015) Substrate-assisted and enzymatic pretransfer editing of nonstandard amino acids by methionyl-tRNA synthetase. Biochemistry 54:5757–5765
Jakubowski H (2012) Quality control in tRNA charging. Wiley Interdiscip Rev RNA 3:295–310
Kermgard E, Yang Z, Michel A-M, Simari R, Wong J, Ibba M, Lazazzera BA et al (2017) Quality control by isoleucyl-tRNA synthetase of bacillus subtilis is required for efficient sporulation. Sci Rep 7:41763
Jakubowski H (2011) Quality control in tRNA charging - editing of homocysteine. Acta Biochim Pol 58:149–163
Jakubowski H (1999) Misacylation of tRNA(Lys) with noncognate amino acids by Lysyl-tRNA synthetase. Biochemistry 38:8088–8093
Dewan V, Reader J, Forsyth KM (2014) In Aminoacyl-tRNA synthetases in biology and medicine Kim S Ed. 344:293–329
Yi J, Cai Z, Qiu H, Lu F, Luo Z, Chen B, Gu Q, Xu J, Zhou H et al (2022) Fragment screening and structural analyses highlight the ATP-assisted ligand. Nucleic Acids Res 50:4755–4768
F CS, P M (2019) Progress and challenges in aminoacyl-tRNA synthetase-based therapeutics. J Biol Chem 294(14):5365–5385. https://doi.org/10.1074/jbc.REV118.002956
Chowdhury S, Nandi N (2022) Conformational fitting of a flexible oligomeric substrate does not explain the enzymatic PET degradation. Phys Chem B 126:620–633
Nakama T, Nureki O, Yokoyama S (2001) Structural basis for the recognition of isoleucyl-adenylate and an antibiotic, mupirocin, by isoleucyl-tRNA synthetase. J Biol Chem 276:47387–47393
Cestari I, Stuart K (2013) Inhibition of isoleucyl-tRNA synthetase as a potential treatment for human African trypanosomiasis. J Biol Chem 288:14256–14263
Johnson RA, Chan AN, Ward RD, McGlade CA, Hatfield BM, Peters JM, Li B et al (2021) Inhibition of isoleucyl-tRNA synthetase by the hybrid antibiotic thiomarinol. J Am Chem Soc 143:12003–12013
Pang L, Nautiyal M, De Graef S, Gadakh B, Zorzini V, Economou A, Strelkov SV, Van Aerschot A, Weeks SD et al (2020) Structural insights into the binding of natural pyrimidine-based inhibitors of class II aminoacyl-tRNA synthetases. ACS Chem Biol 15:407–415
Zeng Y, Kulkarni A, Yang Z, Patil PB, Zhou W, Chi X, Van Lanen S, Chen S et al (2012) Biosynthesis of albomycin δ2 provides a template for assembling siderophore and aminoacyl-tRNA synthetase inhibitor conjugates. ACS Chem Biol 7:1565–1575
Perona JJ, Hadd A (2012) Structural diversity and protein engineering of the aminoacyl-tRNA synthetases. Biochemistry 51:8705–8729
Khan S (2016) Recent advances in the biology and drug targeting of malaria parasite aminoacyl-tRNA synthetases. Malaria J 15
Pang L , Weeks SD, Van Aerschot A et al (2021) Aminoacyl-tRNA synthetases as valuable targets for antimicrobial drug discovery. Int J Mol Sci 22:1422–0067
Molecular Operating Environment (MOE) (2016) Chemical Computing Group Inc.: 1010 Sherbooke St. West, Suite #910, Montreal, QC, Canada, H3A 2R7
Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kale L, Schulten K et al (2005) Scalable molecular dynamics with NAMD. J Comput Chem 26:1781–1802
Vreven T, Morokuma K, Farkas O, Schlegel HB, Frisch MJ et al (2003) Geometry optimization with QM/MM, ONIOM, and other combined methods. I. Microiterations and constraints. J Comp Chem 24:760–769
Cui Q, Guo H, Karplus M (2002) Combining ab initio and density functional theories with semiempirical methods. J Chem Phys 117:5617–5631
Alvarado O, García-Meseguer R, Ruiz-Pernía JJ, Tuñon I, Delgado EJ et al (2021) Mechanistic study of the biosynthesis of R-phenylcarbinol by acetohydroxyacid synthase enzyme using hybrid quantum mechanics/molecular mechanics simulations. Arch Biochem Biophys 701:108807
Aboelnga M, Wetmore SD (2019) Unveiling a single-metal-mediated phosphodiester bond cleavage mechanism for nucleic acids: a multiscale computational investigation of a human DNA repair enzyme. J Am Chem Soc 141:8646–8656
Becke AD (1993) Density-functional thermochemistry. III. The role of exact exchange. J Chem Phys 98:5648
Lee CTY, W T, Parr RG, (1988) Development of the Colle-Salvetti Correlation-Energy Formula into aFunctional of the Electron Density. Phys Rev B 37:785
Frisch MJ, Trucks GW, Schlegel HB, Scuseria GE, Robb MA, Cheeseman JR, Scalmani G, Barone V, Mennucci B, Petersson GA, Nakatsuji H; Caricato, M, Li X, Hratchian HP, Izmaylov AF, Bloino J, Zheng G, Sonnenberg JL, Hada M, Ehara M, Toyota K, Fukuda R, Hasegawa J, Ishida M, Nakajima T, Honda Y, Kitao O, Nakai H, Vreven T, Montgomery Jr JA, Peralta JE, Ogliaro F, Bearpark MJ, Heyd J, Brothers EN, Kudin KN, Staroverov VN, Kobayashi R, Normand J, Raghavachari K, Rendell AP, Burant JC, Iyengar SS, Tomasi J, Cossi M, Rega N, Millam, NJ, Klene M, Knox JE, Cross JB, Bakken V, Adamo C, Jaramillo J, Gomperts R, Stratmann RE, Yazyev O, Austin AJ, Cammi R, Pomelli, Ochterski JW, Martin RL, Morokuma K, Zakrzewski VG, Voth GA, Salvador P, Dannenberg JJ, Dapprich S, Daniels AD, Farkas Ö, Foresman JB, Ortiz JV, Cioslowski J, Fox DJ et al (2009) Gaussian 09 (Revision E.01) Gaussian Inc Wallingford CT
Aboelnga MM, Gauld JW (2022) Comparative QM/MM study on the inhibition mechanism of β-hydroxynorvaline to threonyl-tRNA synthetase. J Mol Graph Model 115:108224
Sklenak S, Yao LS, Cukier RI, Yan HG et al (2004) Catalytic mechanism of yeast cytosine deaminase: an ONIOM computational study. J Am Chem Soc 126:14879–14889
van der Kamp MW, Mulholland AJ (2013) Combined quantum mechanics/molecular mechanics (QM/MM) methods in computational enzymology. Biochemistry 52:2708–2728
Senn HM, Thiel W (2009) QM/MM methods for biomolecular systems. Angew Chem Int Ed Engl 48:1198–1229
Sen A, Gaded V, Jayapal P, Rajaraman G, Anand R et al (2021) Insights into the dual shuttle catalytic mechanism of guanine deaminase. J Phys Chem B 125:8814–8826
Funding
We acknowledge the Natural Science and Engineering Research Council of Canada (NSERC) for financial support and Compute Canada for an award allocation of computational resources.
Author information
Authors and Affiliations
Contributions
Author contribution
M. M. Aboelnga: investigation, methodology, formal analysis, project administration, validation, visualization, and writing—original draft. J. W. Gauld: conceptualization, methodology, project administration, supervision, resources, funding acquisition, and writing.
Corresponding authors
Ethics declarations
Ethical approval
This is not applicable.
Competing interests
The authors declare no competing interests.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Aboelnga, M.M., Gauld, J.W. Establishing a substrate-assisted mechanism for the pre-transfer editing in SerRS and IleRS: a QM/QM investigation. Struct Chem 35, 519–530 (2024). https://doi.org/10.1007/s11224-023-02204-1
Received:
Accepted:
Published:
Issue Date:
DOI: https://doi.org/10.1007/s11224-023-02204-1