Abstract
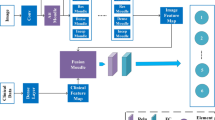
The skin acts as an important barrier between the body and the external environment, playing a vital role as an organ. The application of deep learning in the medical field to solve various health problems has generated increasing interest. The objective of this research is to create a new model using a deep network with multi-feature fusion that enables accurate identification of various skin diseases. This paper proposes a fully fused network (FFN), which includes an improved single block (ISB) and an improved fusion block (IFB) to achieve optimal performance. To implement this, a convolutional neural network-based model for multi-class recognition of skin images has been developed, in which ISB is used to segment diseases in skin images. The IFB module is designed to enhance the effectiveness of the fused network. The model is built and evaluated using our proprietary dataset (Skin_disease_v1) along with publicly available datasets, namely ISIC2016, ISIC2017, and HAM10000. The highest accuracy achieved in experiments was 86% for ISB, 90% for IFB, and 92% for FFN using HAM10000 (for ResNet101V2), HAM10000 (for Resnet50 + ResNet101V2), and HAM10000 (for Resnet50 + ResNet101V2). This shows an accuracy improvement of 9.2%, 13.2%, and 15.2% compared to the state-of-the-art ISB, IFB, and FFN approaches, respectively. Our network outperforms existing networks in terms of performance.









Similar content being viewed by others
Data Availability
The three datasets used in this work are publicly available, and one private dataset is available upon request.
References
Flohr C, Hay R (2021) Putting the burden of skin diseases on the global map[J]. Br J Dermatol 184(2):189–190
Onyema EM, Lilhore UK, Saurabh P, Dalal S, Nwaeze AS, Chi**du AT, ..., Simaiya S (2023). Evaluation of IoT-Enabled hybrid model for genome sequence analysis of patients in healthcare 4.0. Measurement: Sensors, 26, 100679
Edeh MO, Otto EE, Richard-Nnabu NE, Ugboaja SG, Umoke CC, Omachi D (2021) Potential of Internet of Things and semantic web technologies in the health sector. Nigerian J Biotechnol 38(2):73–83
Yang S, Wang H, Chen B (2023) SIBoLS: Robust and Energy-efficient Learning for Spike-based Machine Intelligence in Information Bottleneck Framework. IEEE Transactions on Cognitive and Developmental Systems
Yang S, Chen B (2023) SNIB: improving spike-based machine learning using nonlinear information bottleneck. IEEE Transactions on Systems, Man, and Cybernetics: Systems.
Kharya S, Onyema EM, Zafar A, Wajid MA, Afriyie RK, Swarnkar T, Soni S (2022). Weighted Bayesian belief network: a computational intelligence approach for predictive modeling in clinical datasets. Computational Intelligence and Neuroscience, 2022
Yang S, Chen B (2023) Effective Surrogate Gradient Learning with High-Order Information Bottleneck for Spike-Based Machine Intelligence. IEEE Trans Neural Networks and Learning Systems
Bilgic SA, Cicek D, Demir B (2020) Dermoscopy in differential diagnosis of inflammatory dermatoses and mycosis fungoides[J], Int J Dermatol 59(7): 843–850.d
Yang S, Linares-Barranco B, Chen B (2022) Heterogeneous ensemble-based spike-driven few-shot online learning. Front Neurosci 16:850932
Yang S, Pang Y, Wang H, Lei T, Pan J, Wang J, ** Y (2023) Spike-driven multi-scale learning with hybrid mechanisms of spiking dendrites. Neurocomput 542:126240
Yang Y, **e F, Zhang H, Wang J, Liu J, Zhang Y, Ding H (2023) Skin lesion classification based on two-modal images using a multi-scale fully-shared fusion network. Comput Meth Prog Biomed 229:107315
Hsu BWY, Tseng VS (2022) Hierarchy-aware contrastive learning with late fusion for skin lesion classification. Comput Meth Programs Biomed 216:106666
He X, Wang Y, Zhao S, Chen X (2023) Co-attention fusion network for multimodal skin cancer diagnosis. Pattern Recog 133:108990
Qiu S, Li C, Feng Y, Zuo S, Liang H, Xu A (2023) GFANet: Gated fusion attention network for skin lesion segmentation. Comput Bio Med 155:106462
Feng S, Zhao H, Shi F, Cheng X, Wang M, Ma Y, **ang D, Zhu W, Chen X (2020) CPFNet: context pyramid fusion network for medical image segmentation. IEEE Trans Med Imag 39:3008–3018. https://doi.org/10.1109/TMI.2020.2983721
Tang P, Yan X, Nan Y, **ang S, Krammer S, Lasser T (2022) FusionM4Net: A multi-stage multi-modal learning algorithm for multi-label skin lesion classification. Med Image Analy 76:102307
Wang Y, Haq NF, Cai J, Kalia S, Lui H, Wang ZJ, Lee TK (2022) Multi-channel content-based image retrieval method for skin diseases using similarity network fusion and deep community analysis. Biomed Signal Proc Contr 78:103893
Gairola AK, Kumar V, Sahoo AK (2022) Exploring the strengths of Pre-trained CNN Models with Machine Learning Techniques for Skin Cancer Diagnosis. 2022 IEEE 2nd Mysore Sub Section International Conference (MysuruCon), Mysuru, India. 1-6https://doi.org/10.1109/MysuruCon55714.2022.9972741
Anand V, Gupta S, Koundal D, Singh K (2023) Fusion of U-Net and CNN model for segmentation and classification of skin lesion from dermoscopy images. Expert Syst Appl 213:119230
Gairola AK, Kumar V, Sahoo AK (2023). Deep Learning based Multiple Skin Disease Classification in Indian Territory. In 2023 International Conference on Advancement in Computation & Computer Technologies (InCACCT) (pp. 559-564). IEEE
Khan MA, Sharif M, Akram T, Damaševičius R, Maskeliūnas R (2021) Skin Lesion Segmentation and Multiclass Classification Using Deep Learning Features and Improved Moth Flame Optimization. Diagnostics 11(5):811. https://doi.org/10.3390/diagnostics11050811
Khan MA, Sharif M, Akram T, Bukhari SAC, Nayak RS (2020) a). Developed Newton-Raphson based deep features selection framework for skin lesion recognition. Pattern Recog Lett 129:293–303. https://doi.org/10.1016/j.patrec.2019.11.034
Zhou L, Liang L, Sheng X (2023) GA-Net: Ghost convolution adaptive fusion skin lesion segmentation network. Comput Bio Med 164:107273
Bi L, Feng DD, Fulham M, Kim J (2020) Multi-label classification of multi-modality skin lesion via hyper-connected convolutional neural network. Patt Recog 107:107502
Back S, Lee S, Shin S, Yu Y, Yuk T, Jong S, ..., Lee K (2021) Robust skin disease classification by distilling deep neural network ensemble for the mobile diagnosis of herpes zoster. IEEE Access, 9, 20156-20169.
Tang P, Liang Q, Yan X, **ang S, Zhang D (2020) GP-CNN-DTEL: Global-part CNN model with data-transformed ensemble learning for skin lesion classification. IEEE J Biomed Health Inform 24(10):2870–2882
Gu Y, Ge Z, Bonnington CP, Zhou J (2019) Progressive transfer learning and adversarial domain adaptation for cross-domain skin disease classification. IEEE J Biomed Health Inform 24(5):1379–1393
Wang D, Pang N, Wang Y, Zhao H (2021) Unlabeled skin lesion classification by self-supervised topology clustering network. Biomed Signal Proc Control 66:102428
Al-Masni MA, Kim DH, Kim TS (2020) Multiple skin lesions diagnostics via integrated deep convolutional networks for segmentation and classification. Comput Meth Prog Biomed 190:105351
Author information
Authors and Affiliations
Corresponding author
Ethics declarations
Conflicts of Interest
The authors declare that they have no conflict of interest.
Additional information
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Rights and permissions
Springer Nature or its licensor (e.g. a society or other partner) holds exclusive rights to this article under a publishing agreement with the author(s) or other rightsholder(s); author self-archiving of the accepted manuscript version of this article is solely governed by the terms of such publishing agreement and applicable law.
About this article
Cite this article
Gairola, A.K., Kumar, V., Sahoo, A.K. et al. Multi-feature Fusion Deep Network for Skin Disease Diagnosis. Multimed Tools Appl (2024). https://doi.org/10.1007/s11042-024-18958-7
Received:
Revised:
Accepted:
Published:
DOI: https://doi.org/10.1007/s11042-024-18958-7